- EN - English
- CN - 中文
Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software
利用免疫荧光和Fiji软件定量分析辐照细胞中γH2AX焦点的实验方案
(§Technical contact: Lu_Deng0821@outlook.com) 发布: 2025年08月20日第15卷第16期 DOI: 10.21769/BioProtoc.5421 浏览次数: 2413
评审: Pilar Villacampa AlcubierreAnonymous reviewer(s)
Abstract
Quantification of DNA double-strand breaks (DSBs) is critical for assessing genomic damage and cellular response to stress. γH2AX is a well-established marker for DNA double-strand breaks, but its quantification is often performed manually or semi-quantitatively, lacking standardization and reproducibility. Here, we present a standardized and automated workflow for γH2AX foci quantification in irradiated cells using immunofluorescence and a custom Fiji macro. The protocol includes steps for cell irradiation, immunostaining, image acquisition, and automated foci counting. The protocol is also adaptable to colony-like formations in multi-well plates, extending its utility to clonogenic assays. This protocol enables high-throughput, reproducible quantification of DNA damage with minimal user bias and can be readily implemented in routine laboratory settings.
Key features
• Provides an automated Fiji macro for high-throughput quantification of nuclear γH2AX fluorescence foci with single-nucleus resolution.
• Standardized workflow optimized for reproducibility and cross-sample consistency in DSBs detection.
• Applicable to nuclear fluorescence foci, as well as colony-like structures in multi-well formats for DNA damage and clonogenic assays.
Keywords: γH2AX (γH2AX)Graphical overview
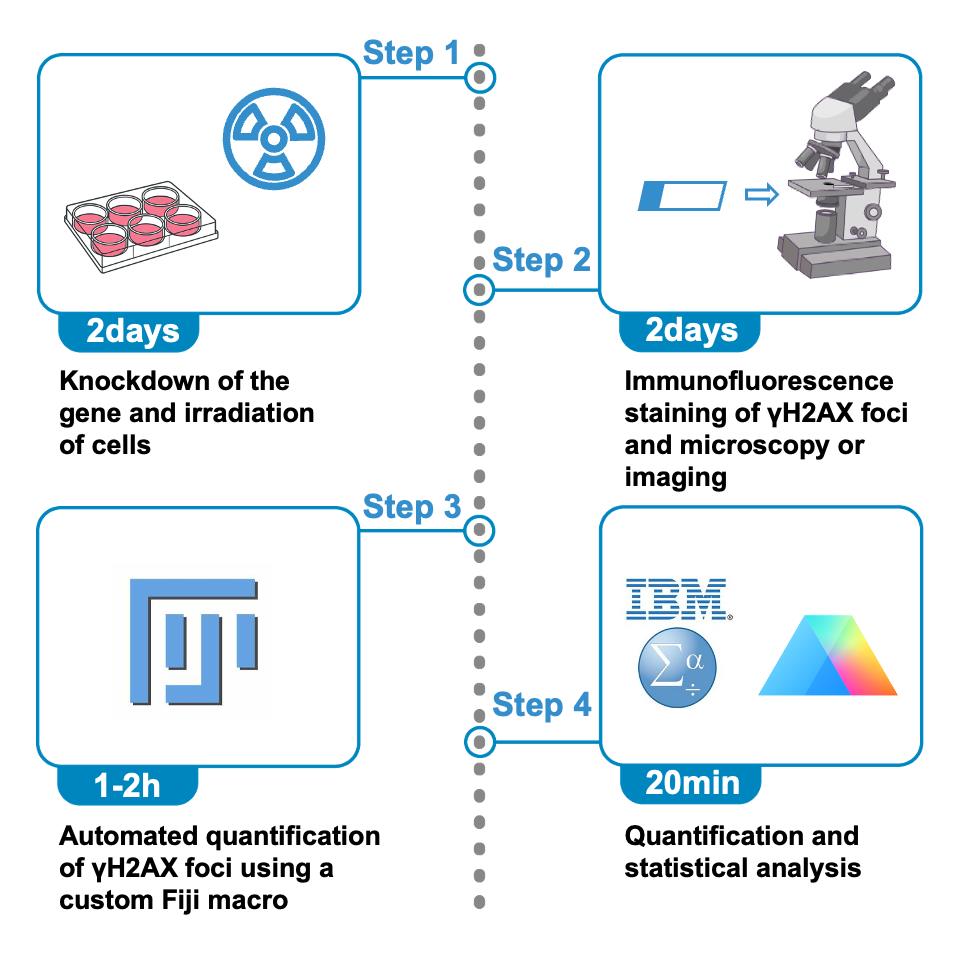
Schematic overview of the protocol for quantifying γH2AX foci in irradiated cells using immunofluorescence and Fiji. The procedure begins with gene knockdown and X-ray irradiation of cultured cells (step 1, ~2 days), followed by immunofluorescence staining of γH2AX foci and image acquisition using fluorescence microscopy (step 2, ~2 days). Automated quantification of γH2AX foci is then performed using a custom Fiji macro (step 3, ~1–2 h). Finally, the resulting data are subjected to statistical analysis and visualization (step 4, ~20 min).
Background
Histone H2AX plays essential roles in nucleosome assembly, chromatin remodeling, and DNA repair [1]. DSBs can be induced by various sources, including ionizing radiation, chemical agents (such as etoposide, doxorubicin, and camptothecin), and endogenous biological processes (such as replication stress). Upon induction of DSBs, H2AX is rapidly phosphorylated at serine 139 to form γH2AX [2]. This phosphorylated histone marks the sites of double-strand breaks and recruits cell cycle checkpoint proteins and DNA repair factors to the damaged regions. Since γH2AX levels rise within minutes after double-strand breaks, it is widely used as a biomarker for DNA damage [1]. Quantitative detection of γH2AX expression can be employed to assess the extent of DNA damage [3].
Currently, the quantification of nuclear fluorescence foci such as γH2AX is predominantly performed using manual or semi-quantitative approaches. Manual counting is labor-intensive and subject to observer bias. Some studies adopt intensity-based or area-based metrics as surrogates for foci number, but these lack spatial resolution and fail to distinguish overlapping or clustered foci. Moreover, many published studies do not explicitly describe how foci are quantified, posing challenges to reproducibility and cross-study comparisons.
To address these limitations, we present an automated, reproducible workflow that performs direct foci counting at the single-nucleus level using a custom Fiji macro. Fiji, a widely adopted open-source platform for biomedical image analysis, supports a wide range of file formats (e.g., TIFF, ND2, CZI) and provides robust tools for fluorescence analysis, morphological quantification, 3D reconstruction, and workflow automation through customizable macros and scripting [4,5]. Our protocol leverages Fiji’s core capabilities while maintaining a user-friendly design that requires little coding experience, making it readily accessible to researchers with diverse technical backgrounds.
This protocol offers a complete and streamlined workflow encompassing sample preparation, immunostaining, image preprocessing, and Fiji macro-based quantification. The approach enables nucleus-by-nucleus quantification of fluorescence foci across experimental groups. It is demonstrated using irradiated C-33A cells, with or without siRNA-mediated knockdown of the target gene, allowing assessment of gene-specific effects on cellular radiosensitivity.
Importantly, beyond γH2AX foci quantification, the macro’s underlying image analysis framework is broadly applicable to other counting tasks based on morphologically similar, segmentable regions of interest (ROIs). For example, it can be used to count colony-like structures in multi-well plate-based assays, including clonogenic assays. This extended applicability underscores the macro’s versatility, enabling a unified and user-friendly workflow for high-throughput quantification of structured, ROI-based image features across diverse experimental settings.
Materials and reagents
Biological materials
1. C-33A cell line (Cell Resource Center, Institute of Basic Medical Sciences, CAMS/PUMC, 1101HUM-PUMC000172)
Reagents
1. LipofectamineTM 2000 (Invitrogen, catalog number: 11668019)
2. GibcoTM basic DMEM, high glucose, pyruvate (Gibco, catalog number: C11995500BT)
3. Fetal bovine serum (FBS) (Gibco, catalog number: A5256701)
4. Pen Strep (Gibco, catalog number: 15140122)
5. Opti-MEMTM (Gibco, catalog number: 31985070)
6. Anti-gamma H2A.X (phospho S139) (mouse) (Abcam, catalog number: ab22551)
7. Goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody, Alexa FluorTM 488 (Thermo Fisher Scientific, catalog number: A-11001)
8. PBS (1×) (Pricella, catalog number: PB180327)
9. Paraformaldehyde, 4% (Solarbio, catalog number: P1110)
10. Triton X-100 (Beyotime, catalog number: P0096)
11. 5% BSA blocking buffer (Solarbio, catalog number: SW3015)
12. Antifade mounting medium with DAPI (Beyotime, catalog number: P0131)
Laboratory supplies
1. Round coverslip (Biosharp, catalog number: BS-18-RC)
2. Size 12 wells, clear polystyrene plate (Corning, catalog number: CLS3513)
3. Parafilm (Sigma-Aldrich, catalog number: P7793)
4. Mendez LASIK forceps (MRP HACO, catalog number: PH2351)
5. Microscope slide (Corning, catalog number: 2947-75X25)
Equipment
1. Upright fluorescence microscope (Leica, catalog number: DM2000)
2. X-ray irradiation (Faxitron, catalog number: MultiRad 225)
Software and datasets
1. Fiji (National Institutes of Health)
2. GraphPad Prism 10 (Dotmatics, Version 10.2.1)
3. IBM SPSS Statistics (IBM Corp., Version 27.0)
Procedure
文章信息
稿件历史记录
提交日期: May 23, 2025
接收日期: Jul 10, 2025
在线发布日期: Aug 5, 2025
出版日期: Aug 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Deng, L., Wang, D. and Wu, L. (2025). Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software. Bio-protocol 15(16): e5421. DOI: 10.21769/BioProtoc.5421.
分类
癌症生物学 > 基因组不稳定性及突变 > 细胞生物学试验 > DNA结构和改变
分子生物学 > DNA > DNA 损伤和修复
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link