- EN - English
- CN - 中文
An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
测定丙酮酸激酶动力学的优化酶偶联分光光度法
发布: 2025年08月20日第15卷第16期 DOI: 10.21769/BioProtoc.5415 浏览次数: 2426
评审: Elena A. OstrakhovitchAnonymous reviewer(s)

相关实验方案

重组人线粒体RNA聚合酶(POLRMT)及启动因子TFAM和TFB2M的表达和纯化
An H. Hsieh [...] Tatiana V. Mishanina
2023年12月05日 2497 阅读
Abstract
Pyruvate kinase M2 (PKM2) is a key glycolytic enzyme that catalyzes the conversion of phosphoenolpyruvate (PEP) to pyruvate, producing ATP in the final step of glycolysis. Unlike other isoforms, PKM2 is uniquely regulated, shifting between active tetramers and less active dimers to balance energy production with biosynthetic demands. This flexibility is exploited in cancer cells to support the Warburg effect and anabolic growth. Additionally, PKM2 can translocate to the nucleus and act as a transcriptional co-activator, influencing gene expression and tumor progression. To facilitate functional studies of PKM2, we present a robust and reproducible protocol for its expression, purification, and enzymatic characterization. PKM2 is expressed in E. coli and purified via Ni-NTA affinity and size-exclusion chromatography to ensure high purity and proper folding. Enzymatic activity is measured using a lactate dehydrogenase (LDH)-coupled assay that tracks NADH oxidation at 340 nm, allowing sensitive kinetic analysis under various conditions, including different PEP concentrations, pH levels, and presence of the allosteric activator fructose-1,6-bisphosphate (FBP). This non-radioactive, high-resolution method is suitable for analyzing PKM2 regulation, post-translational modifications, and mutant variants, as well as for screening potential therapeutic modulators, providing a valuable tool for cancer metabolism research.
Key features
• Enables robust and scalable expression of recombinant wild-type PKM2 in E. coli, yielding protein suitable for biochemical and structural studies.
• Utilizes a non-radioactive, LDH-coupled spectrophotometric assay to accurately measure PKM2 enzymatic activity in real time by monitoring NADH consumption at 340 nm.
• Supports kinetic analysis under physiologically relevant conditions, including variable pH and in the presence or absence of the allosteric activator fructose-1,6-bisphosphate (FBP).
• Suitable for comparative activity profiling of PKM2 variants, mutants, or post-translationally modified forms.
Keywords: PKM2 (PKM2)Graphical overview
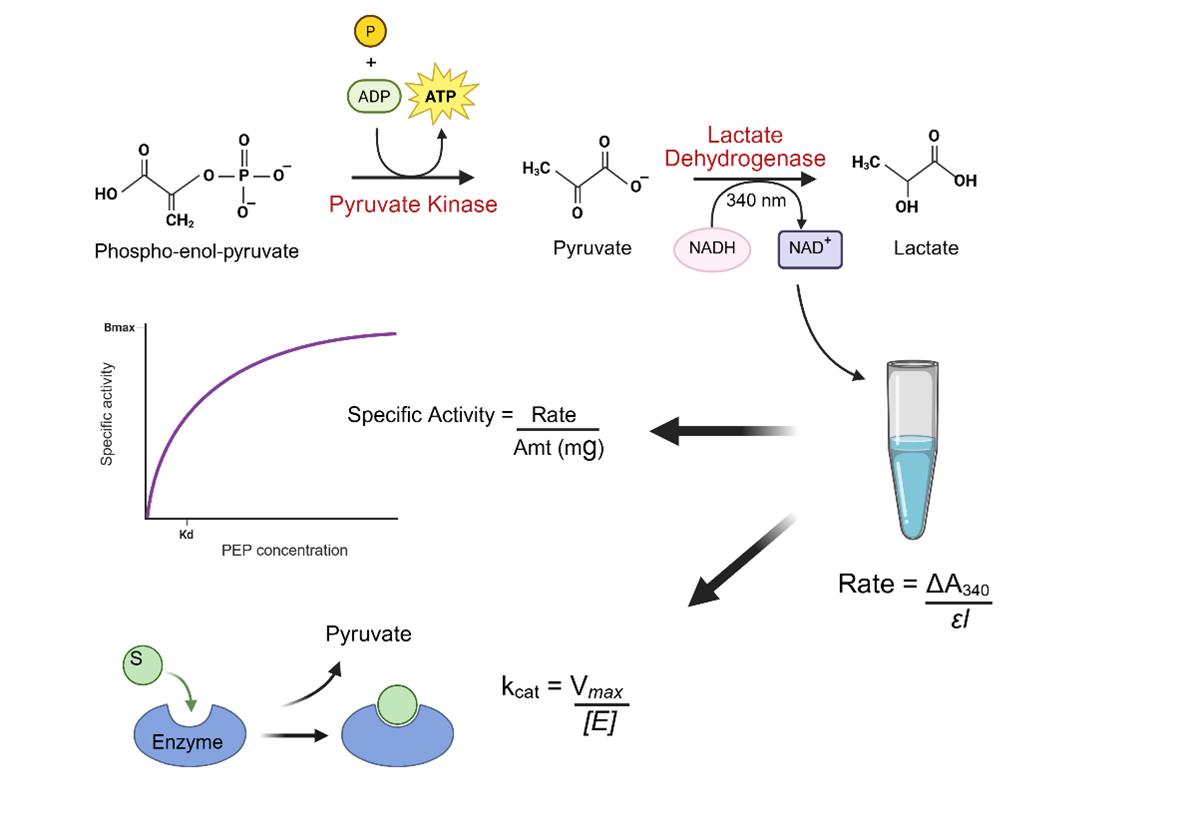
Schematic overview of the coupled enzyme assay for measuring pyruvate kinase activity. The assay monitors pyruvate kinase (PK)-catalyzed conversion of phosphoenolpyruvate (PEP) and ADP to pyruvate and ATP. The pyruvate formed is subsequently reduced to lactate by the lactate dehydrogenase (LDH), which oxidizes NADH to NAD+. The decrease in NADH concentration is measured spectrophotometrically at 340 nm and used to determine enzyme activity. This assay provides a sensitive and quantitative method for analyzing PK activity and enzyme kinetics.
Background
Pyruvate kinase M2 (PKM2) is a key regulatory enzyme in the glycolytic pathway, responsible for catalyzing the final, ATP-generating step in which phosphoenolpyruvate (PEP) is converted to pyruvate [1]. Unlike the other pyruvate kinase isoforms (PKL, PKR, and PKM1), PKM2 is uniquely regulated, capable of existing in multiple oligomeric states—highly active tetramers or less active dimers—allowing it to toggle between promoting energy production and facilitating biosynthetic processes [2,3]. This dynamic regulatory property is particularly relevant in rapidly proliferating and cancerous cells, where PKM2 plays a dual role: maintaining glycolytic flux through the Warburg effect and acting as a nuclear transcriptional co-activator that promotes gene expression linked to cell growth and survival [1,4].
In cancer metabolism, PKM2 serves as a metabolic checkpoint that supports anabolic growth. The less active dimeric form slows pyruvate formation, leading to the accumulation of upstream glycolytic intermediates that are redirected into biosynthetic pathways for nucleotide, amino acid, and lipid synthesis [5]. Meanwhile, nuclear PKM2 modulates gene transcription by interacting with transcription factors such as HIF-1α and β-catenin, thereby linking metabolic status to cellular signaling networks [6,7]. This multifaceted functionality makes PKM2 an attractive therapeutic target, and understanding its biochemical behavior under different conditions is essential for uncovering regulatory mechanisms and discovering modulators.
In recent years, enzyme activity assays have emerged as powerful tools for elucidating the function and inhibition of key enzymes involved in metabolic regulation and disease. Robust enzymatic assays have been successfully developed for a range of enzymes, including D-amino acid oxidase (DAAO), SARS-CoV-2 main protease 3CLpro, and others [8,9]. These tools have proven invaluable for screening small-molecule inhibitors, understanding catalytic mechanisms, and validating potential drug targets. Similarly, there is a critical need for reliable, reproducible assays to study PKM2 activity with high sensitivity and quantitative rigor.
The present protocol addresses this need by offering a streamlined pipeline for the expression, purification, and enzymatic characterization of wild-type PKM2. PKM2 is expressed in Escherichia coli and purified using Ni-NTA affinity chromatography followed by size-exclusion chromatography, resulting in a highly pure, properly folded protein suitable for downstream biochemical and structural studies [10]. A major strength of this protocol lies in its enzymatic activity assay, which employs a lactate dehydrogenase (LDH)-coupled spectrophotometric method to monitor NADH oxidation at 340 nm in real time [11]. This non-radioactive approach allows for continuous kinetic readouts and is both safe and scalable.
The assay is optimized to assess pyruvate kinase activity across a wide range of PEP concentrations, pH values, and in the presence or absence of the allosteric activator fructose-1,6-bisphosphate (FBP). It enables accurate determination of key kinetic parameters such as Km, Vmax, and kcat, and is adaptable to evaluate PKM2 mutants, post-translationally modified forms, and potential inhibitors or activators. Compared to traditional assays using crude lysates or radioactive tracers, this method offers improved specificity, reproducibility, and quantitative resolution. This protocol improves upon existing PK activity assays by offering a higher-resolution kinetic profile, a defined workflow for FBP-stimulated activity, and optimized substrate saturation conditions. Unlike prior generalized methods, this protocol includes a validated, reproducible expression and purification system yielding highly pure PKM2 for consistent biochemical assays. The protocol is also compatible with high-throughput screening platforms and structural techniques like crystallography and cryo-EM, making it a valuable resource for dissecting PKM2 regulation and targeting its activity in cancer and metabolic research.
Materials and reagents
Biological materials
1. pET28a vector containing wild-type PKM2 gene (available from Addgene #25538); store at -20 °C
2. E. coli BL21 (DE3) competent cells (NEB, catalog number: C2527H); store at -80 °C
Reagents
Notes:
1. PEP, NADH, and FBP are light- and temperature-sensitive; handle with care.
2. Buffers should be freshly prepared or stored at 4 °C for up to 2 weeks unless otherwise stated by the vendors.
3. Enzyme and cofactor stocks should be stored in small aliquots to avoid freeze–thaw cycles.
1. Protein ladder (Bio-Rad, catalog number: 1610375); store at -20 °C
2. Coomassie Brilliant Blue R-250** (Sigma-Aldrich, catalog number: B0149); store at room temperature
3. SDS-PAGE Gel kit reagents** (Bio-Rad, catalog number: 1610156)
4. Ammonium persulfate (APS) (catalog number: 1610700)
5. TEMED, 5 mL (catalog number: 1610800) and 50 mL (catalog number: 1610801)
6. Gel buffers: Resolving (catalog number: 1610798) and stacking (catalog number: 1610799)
7. Sample buffers: 4× Laemmli (catalog number: 1610747) and 2× Laemmli (catalog number: 1610737)
8. 2-Mercaptoethanol (β-mercaptoethanol) (catalog number: 1610710)
9. Dithiothreitol (DTT) (catalog numbers: 1610610)
10. Fructose-1,6-bisphosphate (FBP), trisodium salt (Sigma-Aldrich, catalog number: F6803); 100 mM stock, store at -80 °C
11. PEP, sodium salt (Sigma-Aldrich, catalog number: P7127); 100 mM stock, store at -20 °C
12. ADP, disodium salt (Sigma-Aldrich, catalog number: A2754); 100 mM stock, store at -20 °C
13. NADH (β-Nicotinamide adenine dinucleotide, reduced form) (Sigma-Aldrich, catalog number: N7410); 10 mM stock, light-sensitive, store at -20 °C in amber tubes
14. Lactate dehydrogenase (LDH) from rabbit muscle (Sigma-Aldrich, catalog number: L1254); ≥4 U/mL final, aliquots stored at -20 °C
15. MgCl2 (Sigma-Aldrich, catalog number: M8266); 1 M stock stored at 4 °C
16. EDTA disodium salt dihydrate (Sigma-Aldrich, catalog number: E5134); 0.5 M stock stored at room temperature
17. Glycerol (Sigma-Aldrich, catalog number: G5516); store at room temperature
18. Imidazole (Sigma-Aldrich, catalog number: I5513); 1 M stock stored at 4 °C
19. NaCl (Sigma-Aldrich, catalog number: S7653); store at room temperature
20. KCl (Sigma-Aldrich, catalog number: P5405); store at room temperature
21. Tris-HCl, pH 7.5 (Sigma-Aldrich, catalog number: T6066); 1 M stock stored at 4 °C
22. Kanamycin sulfate (Sigma-Aldrich, catalog number: K1377); 50 mg/mL stock stored at -20 °C
23. Terrific Broth (TB) medium (Invitrogen, catalog number: 22711022); store at room temperature
24. Luria–Bertani (LB) medium (Invitrogen, catalog number: 12780052); store at room temperature
25. HEPES, pH 7.5 (Sigma-Aldrich, catalog number: 83264); 1 M stock stored at 4 °C
Solutions
1. Buffers for PKM2 purification (see Recipes)
Buffer A (lysis/binding buffer)
Elution buffer
Storage buffer (for dialysis and SEC)
2. Buffers and stocks for PKM2 activity assay (see Recipes)
Assay buffer
PEP stock
ADP stock
FBP stock
NADH stock
LDH stock
PKM2 enzyme stock
3. Additional recipes (see Recipes)
Luria-Bertani (LB) broth
Terrific Broth (TB) medium
Antibiotic stock (kanamycin)
IPTG stock (1 M)
Tris-HCl buffer (1 M, pH 7.5)
KCl solution (1 M)
Imidazole stock (1 M)
EDTA stock (0.5 M, pH 8.0)
DTT stock (1 M)
Recipes
1. Buffers for PKM2 purification
Buffer A (lysis/binding buffer): 20 mM Tris-HCl, pH 7.5; 100 mM KCl; 5 mM imidazole; 1 mM DTT; 5% glycerol
Elution Buffer: Buffer A + 300 mM Imidazole
Storage buffer (for dialysis and SEC): 20 mM Tris-HCl, pH 7.5; 100 mM KCl; 5% glycerol; 1 mM DTT; 0.25 mM EDTA
2. Buffers and stocks for PKM2 activity assay
Assay buffer: 20 mM Tris-HCl, pH 7.5 (or 7.0/8.0 as needed); 150 mM KCl; 5 mM MgCl2; 0.5 mM NADH; 4–5 U/mL LDH.
PEP stock: 100 mM in water, aliquot and store at -20 °C, dilute freshly before use.
ADP stock: 100 mM in water, adjust pH to 7.0, store at -20 °C.
FBP stock: 100 mM in water; store at -80 °C.
NADH stock: 10 mM in water. This is light-sensitive; aliquot and store at -20 °C.
LDH stock: Prepare as per vendor instructions; dilute to working concentration just before use.
PKM2 enzyme stock: Thaw on ice; avoid repeated freeze–thaw cycles.
3. Additional recipes
Luria-Bertani (LB) Broth: 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl. Add distilled water to 1 L. Adjust pH to 7.0 with NaOH if needed. Sterilize by autoclaving at 121 °C for 15–20 min.
Terrific Broth (TB) medium: 12 g of tryptone, 24 g of yeast extract, 4 mL of glycerol. Dissolve in 900 mL of water. Separately prepare phosphate buffer (2.31 g of KH2PO4 + 12.54 g of KH2PO4 in 100 mL of water). Combine both solutions and adjust volume to 1 L. Sterilize by autoclaving at 121 °C for 15–20 min.
Antibiotic stock (kanamycin): Dissolve kanamycin sulfate in sterile water to make a 50 mg/mL stock solution. Filter sterilize using a 0.22 μm filter. Store aliquots at -20 °C.
IPTG stock (1 M): Dissolve 2.38 g of IPTG in 10 mL of distilled water to obtain a 1 M concentration. Filter sterilize (0.22 μm). Store at -20 °C in small aliquots.
Tris-HCl buffer (1 M, pH 7.5): Dissolve 121.1 g of Tris base in ~800 mL of distilled water. Adjust pH to 7.5 with concentrated HCl. Bring volume to 1 L. Sterilize by autoclaving or filter sterilization, depending on the application.
KCl solution (1 M): Dissolve 74.55 g of KCl in 1 L of distilled water. Sterilize by autoclaving. Store at room temperature or 4 °C.
Imidazole stock (1 M): Dissolve 68.08 g of imidazole in 1 L of distilled water. Adjust pH to 7.5 if needed. Filter sterilize and store at 4 °C.
EDTA stock (0.5 M, pH 8.0): Dissolve 186.1 g of disodium EDTA in ~800 mL of water. Adjust pH to 8.0 using NaOH pellets (EDTA dissolves only at pH 8.0). Bring to 1 L and sterilize by filtration.
DTT stock (1 M): Dissolve 15.4 g of DTT in 100 mL of water. Aliquot and store at -20 °C. DTT is unstable in solution.
Laboratory supplies
1. UV-transparent 96-well plates (Corning, catalog number: 3635); store at room temperature
2. Centrifugal protein concentrators (Amicon Ultra-15, 10–30 kDa MWCO, Millipore Sigma)
3. Dialysis tubing (Thermo Fisher, MWCO 10 kDa, catalog number: 88243)
4. Cryovials (Corning, catalog number: 430661); store at room temperature
5. Multichannel pipettes
6. Superdex 200 Increase 10/300 GL column** (Cytiva, catalog number: 28990944); use with FPLC
7. Ni2+-NTA resin** (Qiagen, catalog number: 30210); store at 4 °C
Equipment
1. Epoch 2 microplate spectrophotometer (BioTek)
2. ÄKTA pure FPLC system (Cytiva)
3. UV-Vis spectrophotometer (e.g., NanoDrop 2000c, Thermo Scientific)
4. Refrigerated centrifuge (swinging bucket and fixed-angle rotor) (Beckman Coulter, model: Avanti J-15R)
5. Sonicator with microtip (Branson, model: Sonifier® SFX150)
6. Temperature-controlled incubator shaker (22–37 °C) (Eppendorf, model: Innova S44i Shaking Incubator)
7. UV-Vis spectrophotometer (e.g., NanoDrop 2000c)
8. SDS-PAGE electrophoresis setup (Bio-Rad)
9. pH meter (METTLER TOLEDO)
10. Autoclave (for media and buffer sterilization)
11. -20 °C and -80 °C freezers
12. Ice bucket or cold room for protein work
13. Dialysis tubing or cassette
Software and datasets
1. GraphPad Prism (v. 10)
2. Microsoft Excel
Procedure
文章信息
稿件历史记录
提交日期: May 18, 2025
接收日期: Jul 10, 2025
在线发布日期: Aug 5, 2025
出版日期: Aug 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Upadhyay, S. (2025). An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics. Bio-protocol 15(16): e5415. DOI: 10.21769/BioProtoc.5415.
分类
生物化学 > 蛋白质 > 活性
生物化学 > 蛋白质 > 定量
分子生物学 > 蛋白质 > 表达
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link