- EN - English
- CN - 中文
An Ex Vivo Protocol to Assess IRE1α-Dependent RNA Cleavage Using Total RNA Isolated from Mouse Tissues
利用小鼠组织总RNA的离体检测IRE1α依赖性RNA切割的实验方案
(*Contributed equally to this work, §aTechnical contact: mamun.phiiuc@gmail.com, §bTechnical contact: syrah76@jbnu.ac.kr) 发布: 2025年08月20日第15卷第16期 DOI: 10.21769/BioProtoc.5414 浏览次数: 1839
评审: Al Borhan BayazidNavnita DuttaAnonymous reviewer(s)

相关实验方案
基于蛋白筛选策略从合成文库中分离抗原特异性纳米抗体:结合 MACS 的酵母展示筛选与 FLI-TRAP 方法
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
2026年01月20日 406 阅读
Abstract
Regulated IRE1-dependent decay (RIDD) is a critical cellular mechanism mediated by the endoplasmic reticulum (ER) stress sensor IRE1α, which cleaves a variety of RNA targets to regulate ER homeostasis. Current in vitro assays to study IRE1α activity largely rely on synthetic or in vitro transcribed RNA substrates, which may not fully replicate the physiological complexities of native RNA molecules. Here, we present a comprehensive protocol to assess IRE1α-dependent RNA cleavage activity using total RNA isolated directly from mouse tissues. This protocol provides a step-by-step guide for tissue collection, RNA isolation, an ex vivo RIDD assay, cDNA synthesis, and subsequent RT-PCR analysis of target mRNA cleavage products. Key reagents include active IRE1α protein, the RIDD-specific inhibitor 4μ8C, and target-specific primers for RIDD-regulated genes such asBloc1s1 and Col6a1. Quantitative assessment is achieved using agarose gel electrophoresis and imaging software. This methodology enables the study of IRE1α's RNA cleavage activity under conditions that closely mimic in vivo environments, providing a more physiologically relevant approach to understanding the role of RIDD in cellular and tissue-specific contexts.
Key features
• Uses total RNA from mouse tissues instead of synthetic RNA to better reflect in vivo conditions.
• Includes RIDD-specific controls such as IRE1α inhibitor (4μ8C) and RNase A to confirm targeted RNA cleavage.
• Combines agarose gel electrophoresis and ImageJ quantification for both qualitative and statistical validation.
• Allows comparative studies of IRE1α activity across multiple mouse tissues in different biological contexts.
Keywords: IRE1α (IRE1α)Graphical overview
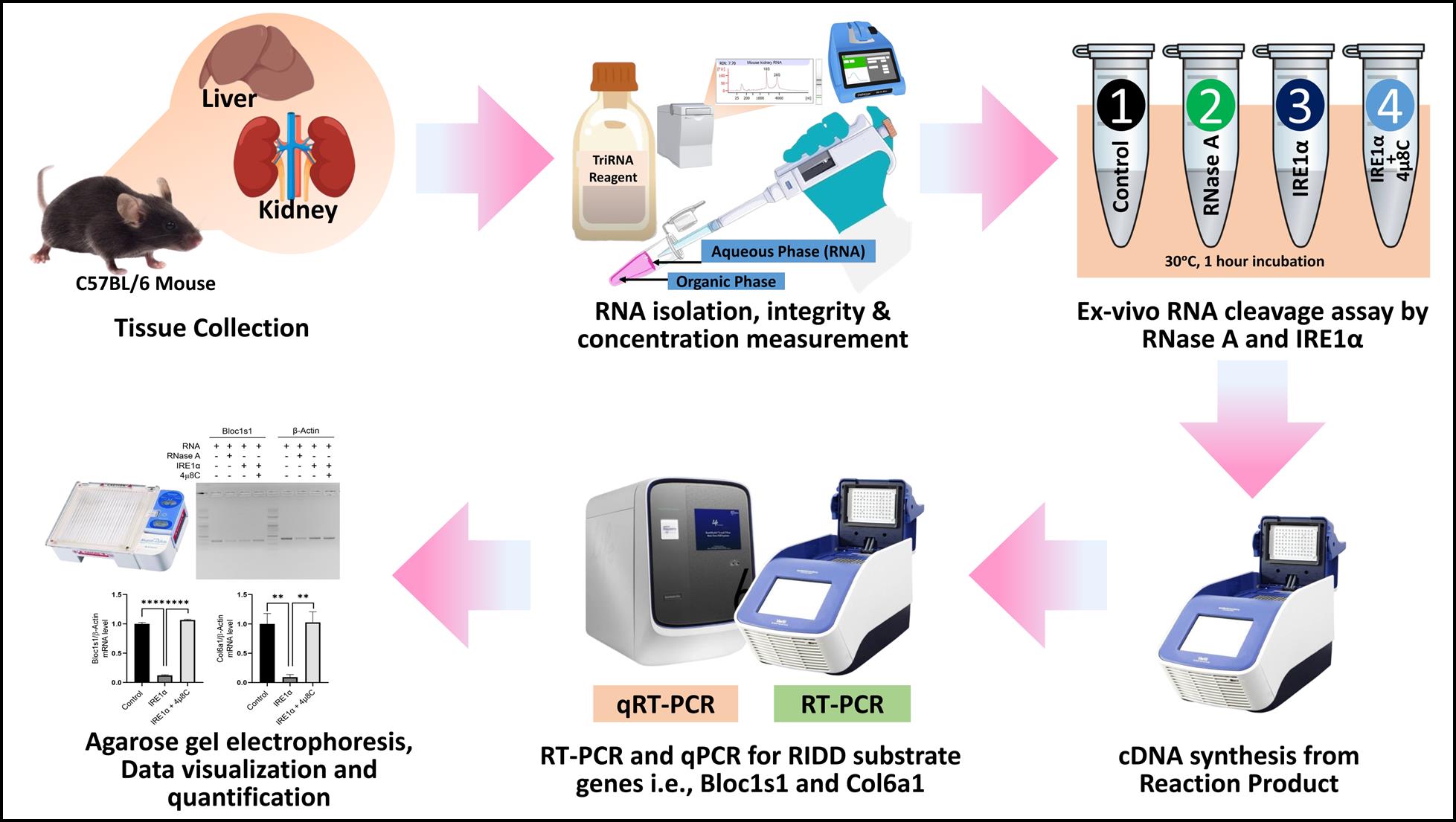
Ex vivo regulated IRE1-dependent decay (RIDD) assay workflow
Background
Regulated IRE1-dependent decay (RIDD) is a cellular mechanism that was first described inDrosophila melanogaster cells, where the endoplasmic reticulum (ER) stress sensor IRE1 was observed to induce the degradation of mRNAs encoding proteins that transit through the ER [1]. Subsequent studies in mammalian cells reported that IRE1α suppressed the expression not only of secretory pathway cargo proteins but also of ER-resident proteins that handle the folding and trafficking of cargo proteins [1].
Under high ER stress, IRE1α acquires endonucleolytic activity against a large array of RNA targets, first identified inD. melanogaster and termed RIDD, including ER-localized mRNAs and non-coding RNAs in mammals [2]. The RIDD pathway was later confirmed in mammalian cells, but it has yet to be fully determined in fungi [3].
The physiological significance of RIDD has been explored in various contexts. RIDD has been shown to regulate IgM and IgG2b expression in B cells, while IgG1 response relies on XBP-1 [4]. RIDD has also been implicated in curtailing immunoglobulin secretion from plasma cells and regulating autophagy in plants by degrading the mRNAs of several negative regulators of autophagy [5,6]. Additionally, RIDD has been suggested to play cytoprotective roles, such as contributing to feedback control on proinsulin expression in pancreatic beta-cells or protecting liver cells from acetaminophen toxicity [7].
Previous studies have established robust in vitro protocols to assess the RNA cleavage activity of IRE1α, predominantly utilizing synthetic RNA substrates [8,9]. These methods often rely on short, specifically designed RNA sequences or in vitro–transcribed RNAs mimicking XBP1 mRNA or validated RIDD targets. Such approaches, while valuable, allow precise kinetic measurements of IRE1α activity and have significantly contributed to understanding its RNase function. However, these methods may oversimplify the complex structural and contextual variations found in native RNA molecules within a cellular or tissue environment. This limitation underscores the need for methodologies that better represent the physiological conditions in which IRE1α operates.
While existing protocols effectively demonstrate IRE1α's RNA cleavage activity using synthetic or specific RNA substrates in vitro, these approaches may not fully capture the physiological context of RNA folding and complexity inherent in native tissues. To address this limitation, we propose a methodology utilizing total RNA isolated directly from various mouse tissues. This approach aims to provide a comprehensive understanding of IRE1α's substrate specificity and activity under conditions that closely mimic in vivo environments. By introducing this innovative perspective, our study seeks to expand the applicability of in vitro RNA cleavage assays, offering a versatile tool to unravel the intricate regulatory roles of IRE1α in diverse biological contexts.
Materials and reagents
Biological materials
1. Collected tissues from C57BL/6 mouse, e.g., kidney and liver
2. PCR primers (Table 1)
Table 1. List of primers
| Gene | Primer | Accession no. | Usage |
| Bloc1s1 | F: 5'-CAAGGAGCCTGCAGGAGAAGA-3' (Tm= 58.4 °C) R: 5'-CCAGGAGGGTGAAGTAAGAGG-3' (Tm = 58.2 °C) | NM_015740.3 | RT-PCR, qRT-PCR |
| Col6a1 | F: 5'-TAGCCGCGATGCAGAAGAG-3' (Tm= 59 °C) R: 5'-TTCCTCGCTCCCCCTCATA-3' (Tm = 58.4 °C) | NM_009933.5 | RT-PCR, qRT-PCR |
| Beta Actin | F: 5'-AGGCCAACCGTGAAAAGATGACC-3' (Tm= 62.7 °C) R: 5'-ACCGCTCGTTGCCAATAGTGATGA-3' (Tm= 62.7 °C) | NM_007393.5 | RT-PCR |
| Beta Actin | F: 5'-GGCTGTATTCCCCTCCATCG-3' (Tm= 59 °C R: 5'-CCAGTTGGTAACAATGCCATGT-3' (Tm= 58.5 °C) | NM_007393.5 | qRT-PCR |
| Stock concentration: 100 µM. Source: Bioneer Corporation, Daejeon, Republic of Korea. | |||
Reagents
A. RNA isolation from different mouse tissues
1. Tri-RNA reagent (Favorgen Biotech Corp., catalog number: FATRR 001) (store at 4 °C; shelf life: 12 months)
2. Chloroform (Sigma-Aldrich, catalog number: C2432) (store at room temperature)
3. Isopropanol (Sigma-Aldrich, catalog number: I9616) (store at room temperature)
4. Ethanol (Sigma-Aldrich, catalog number: E7023) (store at room temperature)
5. UltrapureTM DNase/RNase-free distilled water (Invitrogen, catalog number: 10977015) (store at room temperature)
6. Agilent RNA 6000 Nano kit (Agilent Technologies, catalog number: 5067-1511) (store according to the manufacturer's instructions)
7. 1× PBS (Enzynomics, catalog number: EBP008-1000)
B. RNA cleavage assay of isolated RNA with IRE1α peptide
1. IRE1α protein, active (SignalChem, catalog number: E31-11G) (store at -80 °C)
2. IRE1α inhibitor 4μ8C (MedChemExpress, catalog number: HY-19707) (store at -20 °C)
3. RNase A 100 mg/mL (QIAGEN, catalog number: 1007885) (store at -20 °C; shelf life: 12 months)
4. Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014) (store at room temperature)
5. Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266) (Store at room temperature)
6. Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632) (Store at -20 °C)
7. Glycerol (Sigma-Aldrich, catalog number: G5516) (Store at room temperature)
8. Adenosine 5’-triphosphate (ATP) (Sigma-Aldrich, catalog number: A2383) (Store at -20 °C)
C. cDNA synthesis after RNA cleavage
1. PrimeScriptTM RT Reagent kit (Takara, catalog number: RR037A) (store at -20 °C)
D. Checking mRNA expression of RIDD target mRNA
1. SolgTM 2× Taq PCR Smart mix 1 (SolGent Co. Ltd., catalog number: STD01-M50h) (store at -20 °C)
2. 100 bp DNA ladder (Bioneer Corporation, catalog number: D-1035) (store at -20 °C)
3. UltrapureTM agarose (InvitrogenTM, catalog number: 16500-100) (store at room temperature)
E. qRT-PCR
1. Power SYBRTM Green PCR Master Mix (Applied BiosystemsTM, catalog number: 4367659) (store at -20 °C)
2. 1 M HEPES buffer pH 7.4 (Bio-solution Co., Ltd., catalog number: BH048) (store at room temperature)
3. 50× TAE buffer (Bio-Rad Laboratories, Inc., catalog number: 1610743) (store at room temperature)
4. UltraPureTM ethidium bromide, 10 mg/mL (InvitrogenTM, catalog number: 15585011) (store at room temperature)
Solutions
1. 1 M NaCl (see Recipes)
2. 100 mM ATP (see Recipes)
3. 1 M MgCl2 (see Recipes)
4. 1 M DTT (see Recipes)
5. 5× Reaction buffer (see Recipes)
6. 0.5× TAE buffer (see Recipes)
Recipes
1. 1 M NaCl
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 1 M | 58.44 mg/1 mL |
| UltrapureTM DNase/RNase-free distilled water | - | 1 mL |
| Storage condition: Room temperature; shelf life: 1 month |
2. 100 mM ATP
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| ATP | 100 mM | 55.114 mg/1 mL |
| UltrapureTM DNase/RNase-free distilled water | - | 1 mL |
| Storage condition: -20 °C, avoid repeated freeze-thaw cycles; shelf life: 1 month |
3. 1 M MgCl2
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MgCl2 | 1 M | 95.21 mg/1 mL |
| UltrapureTM DNase/RNase-free distilled water | - | 1 mL |
| Storage condition: Room temperature; shelf life: 1 month |
4. 1 M DTT
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| DTT | 1 M | 15.425 mg/0.1 mL |
| UltrapureTM DNase/RNase-free distilled water | - | 0.1 mL |
| Storage condition: -20 °C, avoid repeated freeze-thaw cycles; shelf life: 1 month |
5. 5× Reaction buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M HEPES pH 7.4 | 100 mM | 10 μL |
| 1 M NaCl | 350 mM | 35 μL |
| 100 mM ATP | 10 mM | 10 μL |
| 1 M MgCl2 | 10 mM | 1 μL |
| 1 M DTT | 25 mM | 2.5 μL |
| Glycerol | 25 % v/v | 25 μL |
| UltrapureTM DNase/RNase-free distilled water | - | 16.5 μL |
| Total volume | 100 μL | |
| *5× reaction buffer is prepared freshly for every experiment. |
6. 0.5× TAE buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 50× TAE buffer | 0.5× | 10 mL |
| Distilled water | - | 990 mL |
| Total volume | 1 L | |
| Storage condition: Room temperature; shelf life: 1 month. |
Laboratory supplies
1. RNase-free 1.5 mL microcentrifuge tubes (Corning Life Sciences Co., Ltd, catalog number: MCT-150-C)
2. Pipette tips 1 mL (Corning Life Sciences Co., Ltd, catalog number: T-1000-B)
3. Pipette tips 200 μL (Corning Life Sciences Co., Ltd, catalog number: T-200-C)
4. Pipette tips 10 μL (Corning Life Sciences Co., Ltd, catalog number: T-300)
5. PCR tubes (Hundaimicro, catalog number: 203017)
6. MicroAmpTM optical 384-well reaction plate with barcode (Applied BiosystemTM, catalog number: 4309849)
7. MicroAmpTM optical adhesive film (Applied BiosystemTM, catalog number: 4311971)
Equipment
1. Veriti 96-well thermal cycler (Applied Biosystem, model: 9902)
2. -80 °C freezer (Nihon Freezer Co., LTD., model: CLN-71UWM)
3. FrescoTM 17 microcentrifuge (Thermo ScientificTM, catalog number: 75002402)
4. Mupid-2plus electrophoresis system (Takara, model: AD110)
5. DeNovix spectrophotometer (DeNovix Inc., model: DS-11+)
6. UltraSlim UV transilluminator (Maestrogen Inc., model: UUV-01)
7. Agilent 2100 bioanalyzer (Agilent Technologies, model: G2939B)
8. QuantStudioTM 6 Flex real-time PCR systems, 384-well, laptop (Applied BiosystemTM, model: 4485691)
Software and datasets
1. ImageJ image processing software (NIH, USA, 1.53e)
2. GraphPad Prism 10 (GraphPad Software, LLC, Boston, USA, 10.2.3)
Procedure
文章信息
稿件历史记录
提交日期: Apr 21, 2025
接收日期: Jul 15, 2025
在线发布日期: Jul 24, 2025
出版日期: Aug 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Rashid, M. M. U., Rah, S., Ko, S. and Chae, H. (2025). An Ex Vivo Protocol to Assess IRE1α-Dependent RNA Cleavage Using Total RNA Isolated from Mouse Tissues. Bio-protocol 15(16): e5414. DOI: 10.21769/BioProtoc.5414.
分类
分子生物学 > RNA > qRT-PCR
生物化学 > 蛋白质 > 相互作用 > Protein-RNA 相互作用
分子生物学 > RNA > 返转录
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link