- EN - English
- CN - 中文
A Fluorescence-Based Flippase Assay to Monitor Lipid Transport by Drs2-Cdc50
基于荧光的Drs2-Cdc50脂质转运活性检测方法
发布: 2025年07月20日第15卷第14期 DOI: 10.21769/BioProtoc.5393 浏览次数: 3225
评审: Anonymous reviewer(s)
Abstract
Flippases, a functionally distinct group of transmembrane proteins that flip lipids from the extracellular or luminal side to the cytosolic side of biological membranes, are key players in many important physiological processes, such as membrane trafficking and cellular signaling. To study the function of these membrane proteins under chemically defined conditions, reconstituting them into artificial vesicles is a crucial and effective approach. There are various methods for protein reconstitution involving different detergents and detergent removal techniques to integrate membrane proteins into artificial vesicles. In this protocol, we describe the reconstitution of the yeast flippase complex Drs2-Cdc50, which translocates phosphatidylserine across membranes of the trans-Golgi network at the expense of ATP hydrolysis. The flippase complex is incorporated into liposomes using a zwitterionic detergent, followed by detergent removal via dialysis—a gentle and effective strategy that helps preserve protein function. To evaluate the activity of the reconstituted flippase complex, two complementary assays are employed: (1) a fluorescence-based quenching assay to measure lipid transport; and (2) an ATPase assay using an ATP-regenerating system to measure ATP hydrolysis. Together, these methods provide a robust platform for analyzing the functional reconstitution of Drs2-Cdc50 in a defined membrane environment.
Key features
• Provides a gentle reconstitution approach via detergent removal by dialysis.
• Measures ATPase activity using an NADH-coupled assay with an ATP-regenerating system.
• Assesses lipid flippase activity with a sodium dithionite-based assay with fluorescent lipid derivatives.
• Provides a well-defined experimental setup for direct characterization of lipid flippases.
Keywords: Large unilamellar vesicles (大单层囊泡)Graphical overview
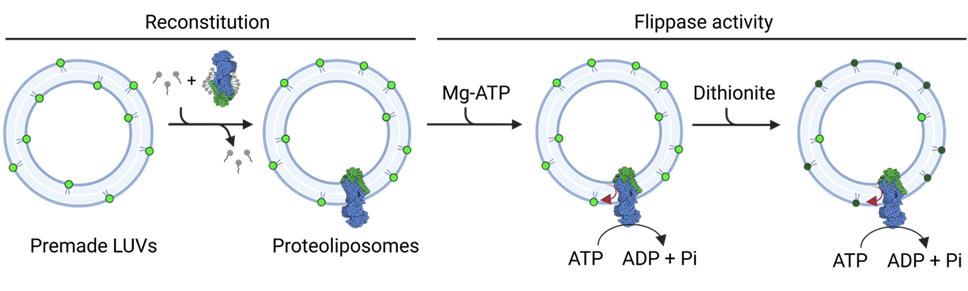
Background
The asymmetrical distribution of lipids across biological membranes is a key characteristic of cells, playing a crucial role in various important physiological processes, including membrane trafficking and cellular signaling [1–5]. Flippases and floppases are essential for the maintenance of phospholipid asymmetry by unidirectional transport of lipids across bilayer leaflets [6]. Eukaryotic flippases, many belonging to the P4-ATPase family, and floppases, which belong to the ABC transporter family, require energy in the form of ATP to move lipids against electrochemical gradients. In contrast, scramblases can transport lipids in both directions without the need of energy, thereby disrupting transbilayer lipid asymmetry [7]. Drs2-Cdc50, a well-studied member of the P4-ATPase family, is primarily localized in the trans-Golgi network of yeast cells. It generates and maintains membrane lipid asymmetry by transporting phosphatidylserine (PS) from the luminal to the cytosolic leaflet of the membrane. This asymmetry is essential for various cellular processes, including vesicle budding and intracellular vesicle trafficking [8]. The role of flippases in these important cellular functions is highlighted by the fact that mutations in human P4-ATPases can lead to pathological conditions in humans. It also makes them attractive drug targets [9].
The molecular study of lipid flippases is challenging due to the complexity of their native membrane environments. Therefore, an important step toward a detailed functional understanding of these proteins is their purification and reconstitution into well-defined, tunable systems such as liposomes. As a first step toward purification, detergent molecules are traditionally used to extract the transmembrane protein from cell membranes, replacing the lipids in lipid–protein interactions [10,11]. This step is critical because, in the absence of lipids, transmembrane proteins are prone to denaturation [12]. To restore their original structure and activity, the purified transmembrane proteins need to be reintroduced into a lipid environment. In traditional reconstitution approaches, the solubilized and purified transmembrane proteins are incorporated into premade large unilamellar vesicles (LUVs) by first destabilizing LUVs with a detergent and then removing the detergent molecules [13]. Typically, non-ionic or zwitterionic, mild detergents are used for vesicle destabilization to preserve the structure and function of the protein. The detergent can then be removed by dialysis, gel filtration, or the use of polystyrene beads that remove the detergent molecules by adsorption [14,15].
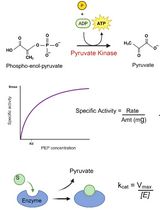
In this protocol, a mild detergent is chosen, and the detergent is removed by dialysis. This approach ensures slow removal, which may help to preserve protein function. However, not all detergents are effectively removed by dialysis. The removal rate depends on the detergent’s critical micelle concentration (CMC), being lower for detergents with low CMCs [14]. As to the study of P4-ATPases flippase activity, many approaches are based on fluorescent nitrobenzoxadiazole (NBD)-labeled lipids [16–18]. These lipids can be incorporated into vesicles either during the preparation of LUVs or during the reconstitution process. In proteoliposomes containing active flippases, the addition of ATP and Mg2+ to the medium will trigger the inward-to-outward movement of NBD lipids. This occurs in vesicles where the catalytic domains of the flippases are facing the outside. To assess flippase activity, the fluorescence of NBD lipids remaining in the outer leaflet is quenched by adding dithionite, a membrane-impermeable reducing agent. Proteoliposomes with functional lipid flippases show increased quenching after incubation with Mg-ATP compared to control vesicles [19]. Alternatively, bovine serum albumin (BSA) back extraction of accessible NBD-lipids can be used to determine lipid flippase activity [19,20]. Another approach to study protein activity is to measure its ability to hydrolyze ATP. This can be done using an ATP-regenerating system that couples ATP hydrolysis to the conversion of NADH to NAD+. This decline of NADH can be tracked by measuring the absorbance at 340 nm [21].
It is important to note that the approach described here relies on ensemble-averaged biochemical assays to assess lipid flippase activity. While these bulk measurements provide valuable quantitative estimates of protein function, they are subject to limitations due to the heterogeneous nature of liposome preparations. Variability in vesicle size, lamellarity, lipid composition, membrane integrity, protein-to-lipid ratio, and the number and orientation of reconstituted proteins can affect data interpretation. For example, only a subset of vesicles may contain functional or correctly oriented proteins, while others may be empty, leaky, or structurally compromised [22–24]. As a result, the activity measured in ensemble assays may not accurately reflect the true behavior of individual protein molecules, leading to underestimation of functional parameters such as transport rates and substrate specificity.
Materials and reagents
Biological materials
The yeast flippase complex Drs2-Cdc50 serves as the exemplary protein in this protocol, with Drs2 carrying an N-terminal biotin acceptor domain (BAD)-tag used for affinity purification. Drs2 was truncated by 104 residues at the N-terminus and 65 residues at the C-terminus (Δ104, Δ65). This truncation is necessary to relieve autoinhibition by the N- and C-termini. The resulting BAD-ΔN104ΔC65Drs2-Cdc50 (from here on referenced as Drs2-Cdc50) was overexpressed from a single co-expression pYeDP60 plasmid in the Saccharomyces cerevisiae strain W303.1b/GAL4 (MAT α, leu2-3, his3-11, trp1-1::TRP1-GAL10-GAL4, ura3-1, ade2-1, canr, cir+) and purified on streptavidin beads [25,26]. The resulting protein stock in elution buffer [50 mM MOPS-Tris, pH 7.0; 100 mM KCl; 10% (v/v) glycerol (Fisher, catalog number: 56-81-5), 0.05% (w/v) n-Dodecyl β-D-maltoside (DDM) (Glycon Biochemicals, catalog number: D97002)] was snap-frozen in liquid N2 and stored at -80 °C.
Note: The flippase complex Drs2-Cdc50 was solubilized using DDM and subsequently stored in DDM to maintain protein stability during storage. For other flippase complexes, optimal solubilization and storage conditions may vary and should be determined empirically.
Reagents
1. 1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Avanti Polar Lipids, catalog number: 850375)
2. 1-Palmitoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoserine (NBD-PS) (Avanti Polar Lipids, catalog number: 810193)
3. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Polar Lipids, catalog number: 850457)
4. 1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) (Avanti Polar Lipids, catalog number: 840457)
5. 3-((3-Cholamidopropyl) dimethylammonio)-1-propansulfonate (CHAPS) (Glycon Biochemicals, catalog number: D99009)
6. 3-(N-morpholino)propanesulfonic acid (MOPS) (Applychem, catalog number: A1076.0100)
7. 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Roth, catalog number: 6763.3)
8. Adenosine 5′-triphosphate disodium salt (ATP) (Roth, catalog number: HN35.3)
9. BeSO4·4H2O (Sigma, catalog number: 14270)
10. Chloroform, ethanol-stabilized and certified for absence of phosgene and HCl (VWR chemicals, catalog number: 22711.260)
11. Deionized water
12. HCl (1 M solution) (VWR chemicals, catalog number: 35328)
13. KOH (1 M solution) (VWR chemicals, catalog number: 35113)
14. Lactate dehydrogenase (LDH, 5 mg/mL) (Sigma, catalog number: 10127230001)
15. L-α-phosphatidylinositol-4-phosphate (PI4P) (Avanti Polar Lipids, catalog number: 840045)
16. Methanol (VWR chemicals, catalog number: 20847.307)
17. MgCl2·6H2O (Sigma, catalog number: M2670)
18. NaCl (Roth, catalog number: 9265.2)
19. NaF (Sigma, catalog number: S1504)
20. NaOH (1 M solution) (VWR chemicals, catalog number: 35256)
21. Nicotinamide adenine dinucleotide disodium salt (NADH) (Sigma, catalog number: 481913)
22. Phosphoenol pyruvate (PEP) (Sigma, catalog number: P0564)
23. Pyruvate kinase (PK, 10 mg/mL) (Sigma, catalog number: P9136-25KU)
24. Sodium dithionite (Fisher, catalog number: CL000239)
25. Tris(hydroxymethyl)aminomethane (Tris) (Roth, catalog number: 4855.1)
26. TritonTM X-100, extra pure (Roth, catalog number: 3051.3)
Solutions
1. Reconstitution buffer (see Recipes)
2. 0.5 M CHAPS (see Recipes)
3. 50 mM MgCl2 (see Recipes)
4. 100 mM PEP (see Recipes)
5. 6.7 mM NADH (see Recipes)
6. 0.5 M ATP (see Recipes)
7. 45 mM beryllium fluoride (see Recipes)
8. Mg-ATP buffer (see Recipes)
9. Na-ATP buffer (see Recipes)
10. 1 M sodium dithionite (see Recipes)
11. 20% (w/v) Triton X-100 (see Recipes)
12. 1 M HEPES-NaOH, pH 4.7 (see Recipes)
13. 1 M NaCl (see Recipes)
14. 1 M MgCl2 (see Recipes)
15. 1 M MOPS-KOH, pH 7 (see Recipes)
16. 1 M Tris-HCl, pH 9 (see Recipes)
17. 0.9 M NaF (see Recipes)
18. 2 M BeSO4 (see Recipes)
Recipes
1. Reconstitution buffer
Adjust the pH to 7.4 with 1 M NaOH before bringing the solution to its final volume with ddH2O. Then, filter sterilize using a 0.22 μm filter.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M HEPES-NaOH, pH 7.4 (see Recipe 12) | 50 mM | 150 mL |
| 1 M NaCl (see Recipe 13) | 100 mM | 300 mL |
| ddH2O | n/a | 2.55 L |
| Total | n/a | 3 L |
2. 0.5 M CHAPS
Store in aliquots at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CHAPS | 0.5 M | 4.61 g |
| ddH2O | n/a | Fill up to 15 mL |
| Total | n/a | 15 mL |
3. 50 mM MgCl2
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M MgCl2 (see Recipe 14) | 50 mM | 750 μL |
| ddH2O | n/a | 14.25 mL |
| Total | n/a | 15 mL |
4. 100 mM PEP
Before adjusting the solution to its final volume with ddH2O, check the pH and adjust it to 7 using 1 M KOH. Store in aliquots at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M MOPS-KOH, pH 7 (see Recipe 15) | 20 mM | 200 μL |
| PEP | 100 mM | 0.19 g |
| ddH2O | n/a | 9.8 mL |
| Total | n/a | 10 mL |
5. 6.7 mM NADH
Before adjusting the solution to its final volume with ddH2O, check the pH and adjust it to 7 using 1 M KOH.
Store in aliquots at -20 or -80 °C and protect from light to prevent degradation.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M MOPS-KOH, pH 7 (see Recipe 15) | 20 mM | 1 mL |
| NADH | 6.7 mM | 0.24 g |
| ddH2O | n/a | 49 mL |
| Total | n/a | 50 mL |
6. 0.5 M ATP
Before adjusting the solution to its final volume with ddH2O, check the pH and adjust it to 7 using 1 M KOH.
Store in aliquots at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| ATP | 0.5 M | 2.25 g |
| ddH2O | n/a | Fill up to 10 mL |
| Total | n/a | 10 mL |
7. 45 mM beryllium fluoride
Caution: Beryllium and its compounds are highly toxic and can cause severe skin burns and eye damage. Please wear appropriate protective gear including gloves, safety goggles, and a lab coat when handling this chemical and perform necessary tasks under a fume hood if possible.
Store in aliquots at -80 °C or prepare a fresh batch before each measurement.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 0.9 M NaF (see Recipe 17) | 0.88 M | 9.775 mL |
| 2 M BeSO4 (see Recipe 18) | 0.045 M | 225 μL |
| Total | n/a | 10 mL |
8. Mg-ATP buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Reconstitution buffer (see Recipe 1) | n/a | 496.55 μL |
| 1 M MgCl2 (see Recipe 14) | 3.3 mM | 1.65 μL |
| 0.5 M ATP (see Recipe 6) | 1.8 mM | 1.8 μL |
| Total | n/a | 500 μL |
9. Na-ATP buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Reconstitution buffer (see Recipe 1) | n/a | 496.55 μL |
| 1 M NaCl (see Recipe 13) | 3.3 mM | 1.65 μL |
| 0.5 M ATP (see Recipe 6) | 1.8 mM | 1.8 μL |
| Total | n/a | 500 μL |
10. 1 M Sodium dithionite
Caution: Sodium hydrosulfide (sodium dithionite) powder is highly toxic if swallowed and can cause severe skin burns and eye damage. Please wear appropriate protective gear, including gloves, safety goggles, and a lab coat, when handling this chemical and prepare the solution under a chemical hood.
Note: Sodium dithionite is unstable in solution; keep the stock on ice and use it within 20–30 min.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M Tris-HCl, pH 9 (see Recipe 16) | 1 M | 500 μL |
| Sodium dithionite | 1 M | 87 mg |
| Total | n/a | 500 μL |
11. 20 % (w/v) Triton X-100
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| TritonTM X-100 | 20% | 20 g |
| ddH2O | n/a | Fill up to 100 mL |
| Total | n/a | 100 mL |
12. 1 M HEPES-NaOH, pH 7.4
Before adjusting the solution to its final volume with ddH2O, check the pH and adjust it to 7.4 using 1 M NaOH.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 1 M | 23.83 g |
| ddH2O | n/a | Fill up to 100 mL |
| Total | n/a | 100 mL |
13. 1 M NaCl
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 1 M | 29.22 g |
| ddH2O | n/a | Fill up to 500 mL |
| Total | n/a | 500 mL |
14. 1 M MgCl2
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MgCl2·6H2O | 1 M | 20.33 g |
| ddH2O | n/a | Fill up to 100 mL |
| Total | n/a | 100 mL |
15. 1 M MOPS-KOH, pH 7
Before adjusting the solution to its final volume with ddH2O, check the pH and adjust it to 7 using 1 M KOH.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MOPS | 1 M | 20.93 g |
| ddH2O | n/a | Fill up to 100 mL |
| Total | n/a | 100 mL |
16. 1 M Tris-HCl, pH 9
Before adjusting the solution to its final volume with ddH2O, check the pH and adjust it to 9 using 1 M HCl.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris | 1 M | 12.11 g |
| ddH2O | n/a | Fill up to 100 mL |
| Total | n/a | 100 mL |
17. 0.9 M NaF
Caution: NaF is toxic and can cause skin irritation. In contact with acids, it develops highly toxic gases. Please wear appropriate protective gear including gloves, safety goggles, and a lab coat when handling this chemical and perform necessary tasks under a fume hood if possible.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaF | 0.9 M | 0.567 g |
| ddH2O | n/a | Fill up to 15 mL |
| Total | n/a | 15 mL |
18. 2 M BeSO4
Caution: Beryllium and its compounds are highly toxic and can cause severe skin burns and eye damage. Please wear appropriate protective gear including gloves, safety goggles, and a lab coat when handling this chemical and perform necessary tasks under a fume hood if possible.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| BeSO4·4H2O | 2 M | 3.543 g |
| ddH2O | n/a | Fill up to 10 mL |
| Total | n/a | 10 mL |
Laboratory supplies
1. Glass cylinder 1 L (e.g., Roth, catalog number: K261.1)
2. Aluminum foil (e.g., VWR chemicals, catalog number: 293-4183)
3. Clamps for dialysis tubes (e.g., Scienova, catalog number: 40330)
4. Dialysis tubing (SERPAVOR, SERVA, catalog number: 44145.01)
5. Filtropur BT25, bottle top filter 0.22 μm (SARSTEDT, catalog number: 83.3940.511)
6. Glass beads, 3 mm (Supelco, catalog number: 1040150500)
7. Glas screw top vials ROTILABO® ND8 (Roth, catalog number: KE30.1)
8. Macro polystyrene cuvettes (SARSTEDT, maximum 4.5 mL volume, catalog number: 67.745)
9. Micro test plate, 96 wells (SARSTEDT, catalog number: 82.1581)
10. Pipette tips 10 μL, 200 μL, and 1,000 μL (SARSTEDT, catalog numbers: 70.3010.305, 70.3030.020, and 70.3050.020)
11. Reaction tubes 1.5 mL (SARSTEDT, catalog number: 72.690.001)
12. Round bottom glass tube (16 × 150 mm, Roth, catalog number: NY90.1)
13. Screw caps ROTILABO® ND8, without borehole, polypropylene (PP), black (Roth, catalog number: KE39.1)
Equipment
1. Analytical balance (e.g., Sartorius Entris-I II, 220 g/0.1 mg; Buch Holm, catalog number: 4669128)
2. Avanti mini extruder set (Avanti Polar Lipids, catalog number: 610000)
a. 1 mL gas-tight syringes (Avanti, no: 610017)
b. 10 mm filter supports (Avanti, no: 610014)
c. 19 mm Nucleopore tracketch membrane with a pore size of 0.2 μm (Schleicher & Schuell)
3. Computer with monitor (e.g., DELL OptiPlex 5090)
4. End-over-end turning device (neoLab, catalog number: 7-0045)
5. Eppendorf Research plus pipettes P2.5, P10, P20, P200, P1000 (Eppendorf, catalog numbers: 3123000012, 3123000020, 3123000039, 3123000055, and 3123000063)
6. Flow cabinet to work with organic solvents
7. Fluorometer (PTI QuantaMaster 800 fluorometers) with integrated FelixGX software, equipped with single cuvette Peltier K-155-C temperature control and magnetic stirrer (Horiba)
8. Freezer (-20 °C) (e.g., Liebherr, model: FNc 4625)
9. Freezer (-80 °C) (e.g., Panasonic, model: VIP plus)
10. Hamilton syringes 25 μL, 50 μL, 100 μL, 500 μL, and 1,000 μL (Hamilton Company, catalog numbers: 20972, 20788, 20790-U, 24539, and 81317)
11. Ice bucket (e.g., Sigma-Aldrich, catalog number: BAM168072002, model: Magic Touch 2)
12. Magnetic stir bar (e.g., Merck, catalog number: HS120548)
13. Magnetic stirrer (e.g., IKA, model: IKAMAG)
14. pH meter (pH-Meter 761 Calimatic, Knick)
15. Refrigerator (4 °C) (e.g., Liebherr, model: FKS 5000)
16. Scissors (e.g., VWR chemicals, catalog number: 233-1280)
17. Microplate reader (ClarioStar, BMG LABTECH)
18. Thermomixer (e.g., Eppendorf, model: Thermomixer 5436)
19. Vacuum pump V-100 with interface I-100 and rotary evaporator Rotavapor® R-100, SJ29/32, V, 220–240V (Buchi, catalog numbers: 11593636, 11593655D, 11100V111, and 11061895)
20. Vortex mixer (e.g., Scientific Industries, model: Vortex Genie 2)
Software and datasets
1. FelixGX software for the control of the PTI QuantaMaster 8000 fluorometer and accessories (Version 4)
2. CLARIOstar software for the control of the CLARIOstar plate reader (Version 5.40 R2)
3. CLARIOstar MARS software for data presentation of plate reader measurements (Version 3.31)
4. Microsoft Excel for data analysis (Version 2505)
Procedure
文章信息
稿件历史记录
提交日期: Apr 1, 2025
接收日期: Jun 16, 2025
在线发布日期: Jul 3, 2025
出版日期: Jul 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Van Der Linden, I. M., Herrera, S. A., Montigny, C., Lenoir, G., Pomorski, T. G. and Uzun, H. D. (2025). A Fluorescence-Based Flippase Assay to Monitor Lipid Transport by Drs2-Cdc50. Bio-protocol 15(14): e5393. DOI: 10.21769/BioProtoc.5393.
分类
生物化学 > 蛋白质 > 活性
生物化学与生物物理学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link