- EN - English
- CN - 中文
Kinase Mobility Shift Assay (KiMSA) for Assessing Protein Kinase A Activity
用于评估蛋白激酶A活性的激酶迁移率变化分析(KiMSA)方法
发布: 2025年07月05日第15卷第13期 DOI: 10.21769/BioProtoc.5366 浏览次数: 2241
评审: Anonymous reviewer(s)
Abstract
The cAMP-dependent protein kinase (PKA) is one of the most extensively distributed kinases among intracellular signal cascades, with a pivotal role in the regulation of various processes, including the capacitation of sperm cells. Traditional assessments of PKA activity rely on the utilization of [γ-32P] ATP and the Kemptide peptide as a substrate. This strategy presents several major drawbacks, including high costs and health risks derived from the manipulation of radioactive isotopes. In this work, we introduce an enhanced non-radioactive assay to quantify PKA activity, termed kinase mobility shift assay (KiMSA), based on the use of a fluorescent-labeled Kemptide (Kemptide-FITC). Once the kinase reaction is terminated, the products can be easily resolved through electrophoresis on an agarose gel and quantified by fluorescence densitometry. We show that KiMSA is suitable for isolated PKA as well as for the enzyme in cell extracts. In addition, it enables quantification of PKA activity during the progression of mouse sperm capacitation. Furthermore, the assay enables monitoring the inhibition of PKA with pharmacological inhibitors in live cells. Therefore, the experimental and optimal assay conditions are set so that KiMSA can be used to assess in vitro as well as in vivo PKA activity in sperm cells. Finally, this method allows for measurement of cAMP concentrations, rendering a versatile technique for the study of cAMP/PKA pathways.
Key features
• KiMSA is a versatile kinase mobility shift assay for measuring PKA activity in sperm physiology, replacing radioactive assays with a fluorescence-labeled substrate for high sensitivity.
• This in vitro assay enables evaluation of activation state, drug effects, or PKA kinetics after purification or mutation.
• This assay measures PKA activity from cell extracts, reflecting pre-lysis activation status and intracellular signaling, though not a true in vivo readout.
• The standard protocol completion time is one day, including sperm preparation, kinase reactions, electrophoresis, and fluorescence quantification.
Keywords: Protein kinase A (PKA) (蛋白激酶A(PKA))Graphical overview
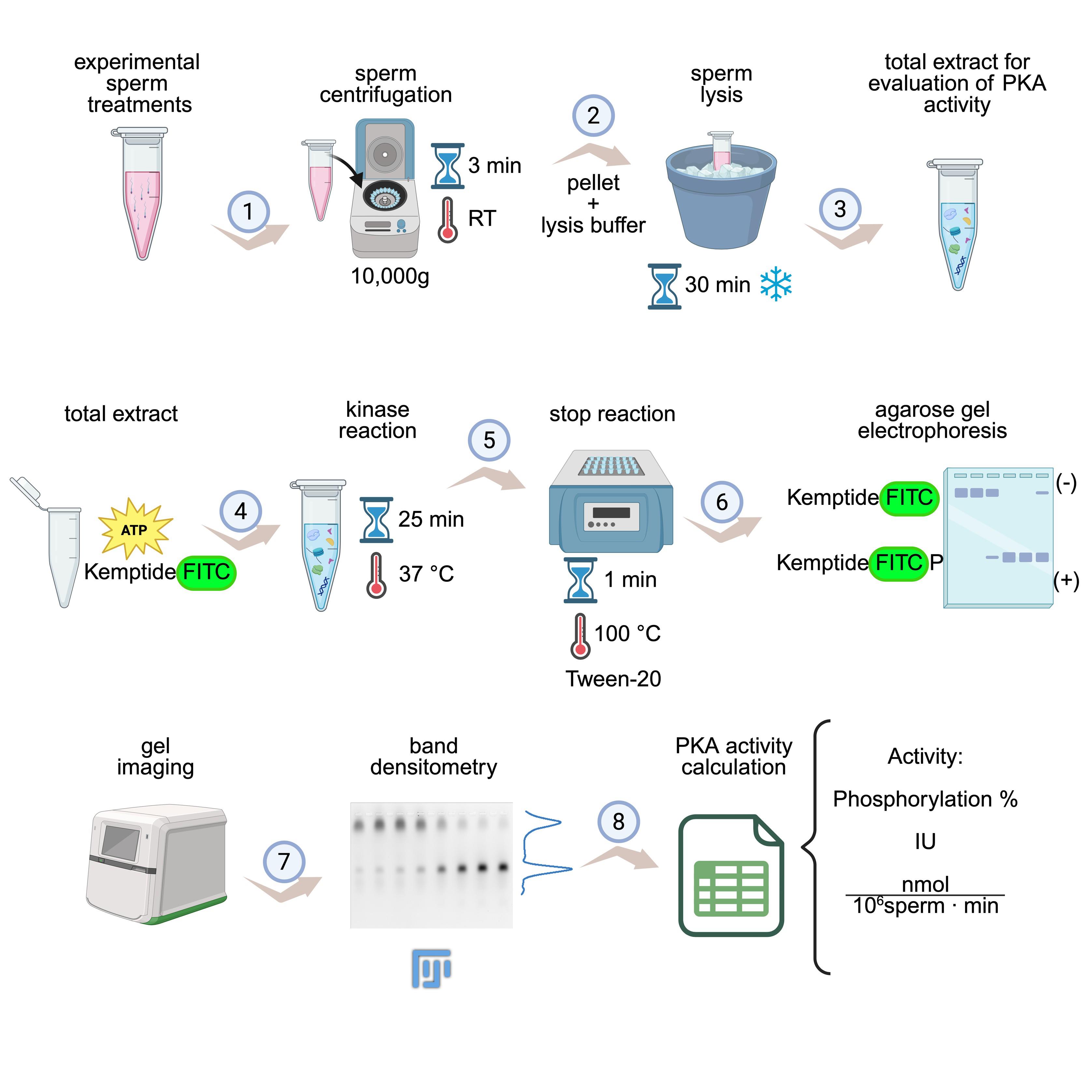
KiMSA workflow summary. Diagram illustrating the steps for obtaining cell extracts for assessing protein kinase A (PKA) activity. Sperm cells are first incubated under the desired experimental conditions (e.g., non-capacitating and capacitating treatments) and then centrifuged at room temperature (RT) for 3 min at 10,000× g (1). Lysis buffer is added to the pellet and incubated for 30 min on ice (2). The required total extract is ready for use (3). Kinase assay is carried out using total extract along with kinase buffer, for 25 min at 37 °C and in the dark (4). Reactions are stopped by quickly placing in ice, adding Tween-20, and incubating at 100 °C for 1 min (5), followed by running on agarose gel electrophoresis (6), gel imaging, and densitometry of phosphorylated and non-phosphorylated Kemptide-FITC signals (7). Finally, PKA activity is calculated as phosphorylation percentage or IU/normalized activity (8). Image modified from the original by Novero et al. [1].
Background
For mammalian sperm to fertilize an oocyte, they first need to undergo a process known as sperm capacitation [2,3], which can be achieved both in vivo during their transit through the female reproductive tract or in vitro by incubating the sperm in a defined capacitating medium [4]. One of the first events during capacitation is the activation of the protein kinase A (PKA), also known as cAMP-dependent kinase A, which phosphorylates proteins on either Ser or Thr residues within the consensus phosphorylation sequence RRXS/T [5]. Early activation of PKA in sperm cells regulates several downstream signaling cascades involved in sperm capacitation [4,6]. In addition, a wide body of literature, which was initiated by the pioneer work of Walsh on phosphorylation related to cAMP-dependent protein kinase activity, indicates that PKA is responsible for phosphorylating a broad array of targets and is considered an essential regulator of many signaling events in somatic cells [7,8]. The PKA holoenzyme is constituted by two regulatory subunits (PKA-R) bound to two catalytic subunits (PKA-C), keeping the enzyme in an inactive state [9–11]. Sperm capacitation can be achieved in vitro upon incubation of sperm in media containing bicarbonate, among other culture medium components, named capacitating media. Bicarbonate stimulates the sperm soluble adenylate cyclase (sAC) [12], triggering an increase in intracellular cAMP. When sperm are incubated under capacitating conditions, the increase of intracellular cAMP induces a conformational change in the regulatory subunits of PKA that releases active PKA-C [5,10]. Since PKA is a central regulator of capacitation and, in turn, of male fertility, investigating its activation is highly relevant in the reproductive field. At present, the predominant approach for assessing direct PKA activity takes advantage of a specific peptide of eight amino acids (LRRASLGK) containing a consensus phosphorylation site for PKA named Kemptide [13,14]. By conducting a catalytic reaction in the presence of [γ-32P]-ATP, the amount of incorporated radioactive phosphate into the Kemptide can be easily correlated to PKA activity [15–17]. However, this methodology has several disadvantages related to the use of 32P, including health risks, high costs, the short half-life of the radioisotope, and the need for trained personnel with specialized facilities. To overcome these drawbacks, we developed a new methodology named KiMSA (Kinase Mobility Shift Assay), where we have labeled the Kemptide using FITC [LRRASLGK-(FITC)], resulting in a compound with a net negative charge at basic pH, which allows it to migrate toward the positive pole (anode) when subjected to agarose gel electrophoresis. Phosphorylation of Kemptide by PKA adds two negative charges to the peptide, resulting in increased migration toward the anode during electrophoresis, allowing the separation of the non-phosphorylated from the phosphorylated version [14,18]. This approach yielded similar results to the standard assay employing [γ32P]-ATP [19] and has been later applied to assess the in vitro activity of PKA purified from pig heart [14]. However, beyond comparable results, KiMSA enables real-time and high-throughput analysis, offering greater sensitivity and quantitative precision across a broader dynamic range. This method also reduces assay variability by directly monitoring substrate binding and phosphorylation events without the need for separation steps, making it more amenable to automation and large-scale screening applications. Collectively, these features position KiMSA as a more efficient, scalable, and safer option for assessing PKA activity in both basic research and drug discovery contexts. In the reproductive field, KiMSA can be easily adapted to measure PKA activity in sperm extracts, a key enzyme during the capacitation process. While previous attempts to use fluorescent-based methods were optimized for purified kinases [18], several issues arise when complex biological samples, such as sperm lysates, are used as the source of the kinase. The assay performance is then hampered by high background fluorescence, peptide aggregation, and debris, which obscure band resolution and reduce sensitivity. The comprehensive protocol provided herein can be used for extracting PKA under conditions that preserve its physiological activation state. Therefore, KiMSA advances the original concept by enabling robust quantification of PKA activity directly in cell extracts. Furthermore, KiMSA accurately reported PKA activation in both non-capacitated and capacitated sperm, truly reflecting the enzyme’s physiological state.
Materials and reagents
Biological materials
1. Sperm from sexually mature C57BL/6 male mice (10–15 weeks old)
Reagents
Note: Unless specified, all reagents are stored at room temperature (RT, 20–25 °C).
1. Bovine serum albumin (BSA) fatty acid-free (Sigma-Aldrich, catalog number: A7906) (4 °C)
2. Adenosine 3-phosphate (ATP) (Sigma-Aldrich, catalog number: A7699) (-20 °C)
3. Cyclic AMP (cAMP) (Sigma-Aldrich, catalog number: A9501) (-20 °C)
4. PhosSTOP phosphatase inhibitor (Roche, catalog number: 4906837001) (4 °C)
5. cOmplete EDTA-free protease inhibitor cocktail (Roche, catalog number: 4693132001) (4 °C)
6. Non-fluorescent Kemptide (sequence LRRASLG) (AnaSpec, catalog number: 22594) (-80 °C)
7. Carboxyl-labeled fluorescent Kemptide (sequence LRRASLGK-FITC, termed Kemptide-FITC) (-80 °C) (Biomatik, catalog number: 1036600)
Note: Additional information on Kemptide-FITC is as follows:
Origin: Synthesized by Biomatik. Sequence: N-LRRASLGK-C-(FITC). MW: 1,289.49 Da. Salt form: trifluoroacetate (TFA Salt). Solubility in water: 1 mg/mL. Purity (HPLC): 96.01%. FITC = fluorescein isothiocyanate. Weigh 1 mg of powder and dissolve it in 1 mL of water (do not weigh less than the resolution limit of the scale). Always keep in the dark to avoid quenching of FITC.
8. Tris(hydroxymethyl)aminomethane (Tris base) (Cicarelli, catalog number: 1131214)
9. Triton X-100 (Neo Lab, catalog number: 01685)
10. NaCl (Cicarelli, catalog number: 750)
11. MgCl2 (Sigma-Aldrich, catalog number: M8266)
12. HEPES (Sigma-Aldrich, catalog number: H3375)
13. KCl (Sigma-Aldrich, catalog number: S-7653)
14. Sodium pyruvate (Sigma-Aldrich, catalog number: P-4562) (4 °C)
15. CaCl2·2H2O (Sigma-Aldrich, catalog number: C-7902)
16. KH2PO4 (Sigma-Aldrich, catalog number: P-5655)
17. MgSO4·7H2O (Sigma-Aldrich, catalog number: 63138)
18. Glucose (Sigma-Aldrich, catalog number: G-6152)
19. NaOH (Biopack, catalog number: 1641.08)
20. Dimethyl-sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D2650)
21. Dithiothreitol (DTT) (BioBasic, catalog number: DB0058) (-20 °C)
22. NaHCO3 (Sigma-Aldrich, catalog number: S6014)
23. 90% Glycerol (Cicarelli, catalog number: 708110)
24. Bromophenol blue (Sigma, catalog number: B-5525)
25. Tween-20 (Sigma-Aldrich, catalog number: SLCL7671)
26. Agarose LE (Productos Bio-Lógicos PB-L, catalog number: IB0501)
27. Source of PKA: recombinant PKA-C (Addgene plasmid, catalog number: 14921, expressed in E. coli BL21 and purified with Ni-NTA resin) or sperm extract (detailed here)
Solutions
1. H-TYH (non-capacitating media) (see Recipes)
2. CAP 2× (see Recipes)
3. 1.5× Triton lysis buffer (incomplete) (see Recipes)
4. 1.5× Triton lysis buffer (complete) (see Recipes)
5. 5× kinase reaction buffer (incomplete) (see Recipes)
6. 5× kinase reaction buffer (complete) (see Recipes)
7. 2.5× FITC-kinase reaction buffer (see Recipes)
8. Agarose loading buffer 10× (see Recipes)
9. Agarose running buffer 1× (see Recipes)
10. 1% (w/v) agarose LE (see Recipes)
Recipes
1. H-TYH (non-capacitating media)
| Reagent | Final concentration (mM) | Quantity for 25 mL (mg) |
|---|---|---|
| NaCl | 119.3 | 174.3 |
| KCl | 4.7 | 8.8 |
| CaCl2 | 1.71 | 6.3 |
| KH2PO4 | 1.2 | 4.1 |
| MgSO4 | 1.2 | 7.4 |
| HEPES (pH 7.2–7.4) | 20 | 119.2 |
| Glucose | 5.4 | 24.8 |
| Sodium pyruvate | 0.8 | 2.2 |
Dissolve all reagents in Mili-Q water (except for glucose and sodium pyruvate) to a final volume of 25 mL. Sterilize by filtration using a 0.45 μM filter and store at 4 °C for up to 1 month. On the day of the experiment, add 2.2 mg of sodium pyruvate and 24.8 mg of glucose. Then, set aside the volume needed to prepare the capacitating medium, depending on experimental conditions (see Recipe 2). Bring the remaining non-capacitating medium to a final pH of 7.2–7.4 with freshly prepared 5 M NaOH.
2. CAP 2×
| Reagent | 2× concentration | Quantity for 5 mL |
| NaHCO3 | 40 mM | 16.8 mg |
| BSA | 10 mg/mL | 50 mg |
| H-TYH media (Recipe 1) | - | 5 mL |
Add NaHCO3 and BSA to the H-TYH media set aside before adjusting the pH to make CAP 2×. Adjust the CAP 2× pH to 7.2–7.4 right before using, as the pH rises with time. To prepare sperm capacitating condition, add one part of CAP 2× to one part of cells + H-TYH.
3. 1.5× Triton lysis buffer (incomplete)
| Reagent | Final concentration | Quantity for 5 mL (μL) |
|---|---|---|
| 1 M Tris (pH 7.4) | 75 mM | 465.8 |
| 3 M NaCl | 225 mM | 465.8 |
| 100% Triton X-100 | 1.5% | 93.1 |
| Distilled water (dH2O) | 3975.3 |
Mix Tris, NaCl, and Triton X-100 and aliquot in volumes of 402.5 μL. Store aliquots at -20 °C. One aliquot is enough to lysate approximately five cell pellets.
4. 1.5× Triton lysis buffer (complete)
| Reagent | Final concentration | Quantity for 500 μL (µL) |
|---|---|---|
| 50× protease inhibitor cocktail ROCHE | 1.5× | 15 |
| 10× PhosSTOP ROCHE phosphatase inhibitor | 1.5× | 75 |
| 1 M DTT | 15 mM | 7.5 |
| 1.5× Triton lysis buffer (incomplete) (Recipe 3) | - | 402.5 |
To one aliquot of incomplete 1.5× Triton lysis buffer, add protease inhibitor cocktail, PhosSTOP, and DTT, and keep on ice. DTT is stable for up to 3 years when refrigerated at 2–8 °C. DTT is hygroscopic, has sensitivity to heat, is incompatible with strong oxidizing agents, and decomposes at pH >7.
It is mandatory to use the EDTA-free version of the EDTA-free protease inhibitor cocktail ROCHE 50× because if a chelating agent is present in the kinase reaction medium, it will interfere with the reaction.
PhosSTOP (phosphatase inhibitor cocktail) can be replaced with 15 mM NaF and 100 mM sodium orthovanadate.
5. 5× kinase reaction buffer (incomplete)
| Reagent | Final 5× concentration (mM) | Quantity for 3 mL (µL) |
|---|---|---|
| 1 M Tris (pH 7.4) | 250 | 1363.6 |
| 1 M MgCl2 | 50 | 272.2 |
| dH2O | - | 1363.6 |
Mix Tris pH 7.4 and MgCl2 and bring to a final volume of 3 mL, then separate into 27.5 μL aliquots and store at -20 °C.
Magnesium acts as a mandatory cofactor for PKA catalysis. On the day of the experiment, each aliquot will be supplemented up to a final volume of 50 μL (allowing for 9–10 reaction tubes).
6. 5× kinase reaction buffer (complete)
| Reagents | Final 5× concentration | Quantity for 50 μL (µL) |
|---|---|---|
| 10 mM ATP (dH2O) | 1 mM | 5 |
| 1 M DTT | 50 mM | 2.5 |
| 50× protease inhibitor Cocktail ROCHE | 5× | 5 |
| 25× PhosSTOP ROCHE | 5× | 10 |
| 5× kinase reaction buffer (incomplete) (Recipe 5) | 5× | 27.5 |
To one aliquot of incomplete 5× kinase reaction buffer, add ATP, protease inhibitor cocktail, PhosSTOP, and DTT. Keep on ice.
7. 2.5× FITC-kinase reaction buffer
| Reagents | Final Concentration | Quantity for 100 μL (µL) |
|---|---|---|
| 5× kinase reaction buffer (complete) (Recipe 6) | 50 | |
| 1 mg/mL Kemptide-FITC | 7.75 pmol/μL* | 10 |
| 3.9 mg/mL Kemptide | 7.75 pmol/μL* | 1.5 |
| dH2O | - | 38.5 |
To a complete 5× kinase reaction buffer aliquot, add Kemptide and Kemptide-FITC. Keep on ice and protected from light to avoid quenching of the fluorophore.
*Note that 1 µg of Kemptide-FITC + 1 µg of Kemptide (not fluorophore-tagged) will be added per reaction tube. 1 µg of either version of the Kemptide is equivalent to 0.775 nmol.
8. Agarose loading buffer 10×
| Reagents | Final 10× concentration | Quantity for 1 mL (µL) |
|---|---|---|
| 85%–89% Glycerol | 63%–66% (v/v) | 740 |
| 5% bromophenol blue (dH2O) | 0.8% (w/v) | 160 |
| 100% Tween-20 | 5% (v/v) | 50 |
| dH2O | - | 50 |
Mix glycerol, bromophenol blue, and Tween-20 and complete to a final volume of 1 mL with dH2O. Aliquot and store at -20 °C.
9. Agarose running buffer 1×
| Reagents | Final concentration | Quantity for 1 L (mL) |
|---|---|---|
| 1.5 M Tris | 50 mM | 33 |
| dH2O | - | 967 |
Adjust pH to 10.0 with 5 M NaOH before completing the volume with water.
This buffer will be used both to run the electrophoresis and to dilute the agarose, so prepare at least 300 mL of running buffer (250 mL will be needed to fill the electrophoresis tank and 50 mL to prepare a small agarose gel of 7 × 10 cm).
10. 1% (w/v) agarose LE
| Reagents | Final concentration | Quantity for 100 mL |
|---|---|---|
| Agarose | 1% (w/v) | 1 g |
| Tris-HCl 50 mM, pH 10.0 | - | 100 mL |
Heat the solution until the agarose is completely dissolved. Allow the mixture to cool to approximately 60 °C before pouring the solution into the gel tray. Add the adequate gel comb according to the sample number. Prepare on the day of use.
Laboratory supplies
1. 0.45 μM filter (Jet Biofil, catalog number: FCA406013)
2. Sterile syringes (Neojet, catalog number: 12238)
3. 15 mL conical tubes (Tarson, catalog number: 546121-RK)
4. 50 mL conical tubes (Tarson, catalog number: 546041-RK)
5. 10 μL pipette tips (Tarson, catalog number: 521000)
6. 200 μL pipette tips (Tarson, catalog number: 521010Y)
7. 1,000 μL pipette tips (Tarson, catalog number: 521016B)
8. 2 mL Eppendorf tubes (Tarson, catalog number: 500020N)
9. 1.5 mL Eppendorf Tubes (Tarson, catalog number: 500010N)
10. 0.6 mL Eppendorf Tubes (Tarson, catalog number: 500000N)
Equipment
1. Neubauer counting chamber (Marienfeld, catalog number: 0640010)
2. Surgery scissors (Merck, catalog number: Z265977)
3. Forceps (Merck, catalog number: F4142)
4. Vortex mixer (DLab, catalog number: 8031102000)
5. Centrifuge (Eppendorf MiniSpin, catalog number: 5452000010 and Eppendorf 5418 R, catalog number: 5401000013)
6. Water bath (37 °C) (Vicking, catalog number: 1002)
7. Heating block (100 °C) (ThermoFisher, catalog number: 88871001)
8. Agarose electrophoresis equipment (Bio-Rad gel casting system, catalog number: 1704481; Bio-Rad Basic Power supply, catalog number: 1645050)
9. Freezer (-20 °C)
10. Refrigerator (2–8 °C)
11. Micropipettes (Gilson; P2, catalog number: F144054M, P10, catalog number: 144055M, P20, catalog number: F144056M, P200, catalog number: F144058M, P1000, catalog number: F144059M)
Software and datasets
1. Microsoft® Office Excel (Microsoft Office 365), or similar
2. FIJI (Fiji is just ImageJ) (Open source, imagej.net, 2.16.0)
3. Typhoon FLA 7000 software under the Fluorescence mode or equivalent fluorescence imager
Procedure
文章信息
稿件历史记录
提交日期: Apr 8, 2025
接收日期: Jun 4, 2025
在线发布日期: Jun 17, 2025
出版日期: Jul 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Novero, A. G., Steeman, T. J., Curcio, C., Buccolini, L., Binolfi, A., Krapf, D., Buffone, M. G., Krapf, D. and Stival, C. (2025). Kinase Mobility Shift Assay (KiMSA) for Assessing Protein Kinase A Activity. Bio-protocol 15(13): e5366. DOI: 10.21769/BioProtoc.5366.
分类
生物化学 > 蛋白质 > 活性
分子生物学 > 蛋白质 > 活性
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link















