- EN - English
- CN - 中文
High-Throughput Indirect Monitoring of TORC1 Activation Using the pTOMAN-G Plasmid in Yeast
基于酵母pTOMAN-G质粒的TORC1活化高通量间接监测方法
(§Technical contact: melissa.gomez.r@usach.cl) 发布: 2025年06月20日第15卷第12期 DOI: 10.21769/BioProtoc.5356 浏览次数: 1601
评审: Ritu GuptaThibaud T. RenaultIsmail Tahmaz

相关实验方案
利用靶向GFP或mCherry的纳米抗体在出芽酵母中一步生成AlissAID条件性敲降菌株
Yoshitaka Ogawa [...] Takumi Kamura
2024年06月20日 2049 阅读
Abstract
The target of rapamycin complex 1 (TORC1) is a highly conserved protein complex whose primary function is to link nutrient availability to cell growth in eukaryotes, particularly nitrogen sources. It was originally identified during the screening of Saccharomyces cerevisiae strains resistant to rapamycin treatment. For its part, S. cerevisiae is well known for being a key model organism in biological research and an essential microorganism for the fermentation of food and beverages. This yeast is widely distributed in nature, with domesticated and wild strains existing. However, little is known about what effects domestication has had on its different phenotypes; for example, how nitrogen sources are sensed for TORC1 activation and what impact domestication has had on TORC1 activation are questions that still have no complete answer. To study the genetic basis of TORC1 activation associated with domestication through approaches such as quantitative trait loci (QTL) mapping or genome-wide association studies (GWAS), and more generally for any study requiring TORC1 activity as a readout for a large number of individuals, it is necessary to have a high-throughput methodology that allows monitoring the activation of this pathway in numerous yeast strains. In this context, the present protocol was designed to assess phenotypical differences in TORC1 activation using a new reporter plasmid, the pTOMAN-G plasmid, specifically designed to monitor TORC1 activation. As a proof of concept, this methodology allowed phenotyping a large population of yeast strains derived from the 1002 Yeast Genomes Project, the most complete catalog of genetic variation in yeasts. This protocol proved to be an efficient alternative to assess TORC1 pathway activation compared to techniques based on immunoblot detection, which, although effective, are considerably more laborious. Briefly, the protocol involves the design and construction of the pTOMAN-G plasmid, which carries a construct containing the firefly luciferase gene (Luc) under the control of the TORC1-regulated RPL26A gene promoter (PRPL26A). The protocol then details the process for selecting subgroups of yeasts based on their ability to grow under nutrient-limited conditions, using proline as the sole nitrogen source. These yeasts are then transformed with the TOMAN-G plasmid, using two alternative transformation methods. Finally, those yeasts that emit luminescence are selected, whose phenotype for TORC1 activation is measured by a nitrogen-upshift experiment in microculture. This approach, using the pTOMAN-G plasmid, offers a rapid and consistent method for assessing TORC1 signaling pathway activation in a large number of yeast strains, highlighting its usefulness to study the activation of the TORC1 pathway and the domestication process associated with it. In the future, a redesign of the plasmid could extend its use as a reporter tool to monitor the activation of the TORC1 pathway, or other pathways, in other yeast species.
Key features
• This protocol is specifically optimized for assessing TORC1 activation in microculture through nitrogen upshift experiments, enabling efficient high-throughput screening of numerous yeast strains.
• This protocol is an improved version of the method applied by Kessi-Pérez et al. [1], being less time-consuming and ensuring better comparability between strains.
• This protocol is only applicable to S. cerevisiae, although a redesign of the pTOMAN-G plasmid could extend its use to other yeast species.
Keywords: Luminescence (发光检测)Graphical overview
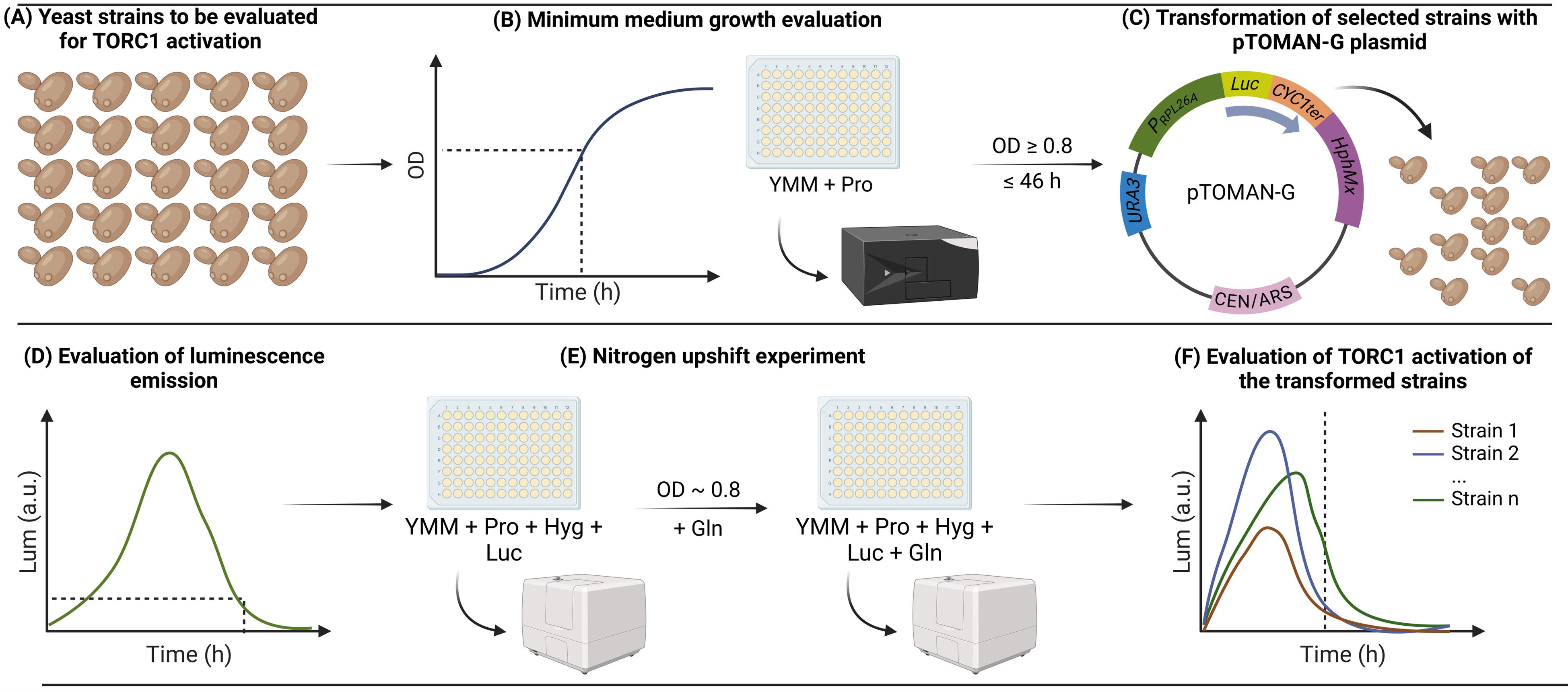
Overview of the method for high-throughput indirect monitoring of TORC1 activation using the pTOMAN-G plasmid in yeast. (A) Yeast strains to be evaluated for TORC1 activation are (B) grown in a minimum medium containing proline as the sole nitrogen source (YMM+Pro), and strains capable of growing to at least an optical density at 600 nm (OD600) of 0.8 in 46 h are (C) transformed with the pTOMAN-G reporter plasmid. Then, (D) the capacity of emitting luminescence by the transformed strains is evaluated, and strains that correctly emit luminescence are (E) evaluated by nitrogen upshift experiments in microculture. Finally, (F) the luminescence curves obtained after the nitrogen pulse are analyzed for every strain evaluated as a readout of TORC1 activation.
Background
Target of rapamycin complex 1 (TORC1) is a highly conserved protein complex whose primary function is to link nutrient availability to cell growth in eukaryotes, particularly nitrogen sources (such as ammonium and amino acids). It was identified during the screening of Saccharomyces cerevisiae strains resistant to rapamycin treatment [2], and since then, this yeast has been fundamental in most of the key advances in understanding the TORC1 signaling pathway [3]. S. cerevisiae, a yeast species well known for being a key model organism in biological research and an essential microorganism for the fermentation of food and beverages, is found in diverse ecological niches and is proposed to be a neutral nomad [4], able to thrive in a wide range of environments: these include natural ecosystems as well as those shaped by human activities, such as the fermentation of grapes for wine production [4]. Consequently, both wild and domesticated strains of this yeast species can be found in different ecological niches. On the other hand, little is known about the genotypic and phenotypic effects associated with the domestication of S. cerevisiae. In recent years, several studies have focused on studying the impact of domestication processes at both phenotypic and genetic levels [5–10], including TORC1 activation [11]. However, how nitrogen sources are sensed for TORC1 activation and what impact domestication has had on TORC1 activation are questions that still have no complete answer.
To study the genetic basis of TORC1 activation associated with domestication through approaches such as quantitative trait loci (QTL) mapping or genome-wide association studies (GWAS), and more generally for any study requiring TORC1 activity as a readout for a large number of individuals (e.g., library screening), it is necessary to have a high-throughput methodology that allows monitoring the activation of this pathway in numerous yeast strains. However, the two most widely used methods to study TORC1 activation, which use phosphorylation of the Sch9 kinase [12] or of the ribosomal protein S6 (Rps6) [13,14] as readouts, are both based on immunoblot detection, which makes them laborious and time-consuming and therefore difficult to use for the analysis of a large number of yeast strains. In this context, in a previous work, we generated a method for high-throughput indirect monitoring of TORC1 activation in microculture through nitrogen upshift experiments (in which yeasts are grown in a medium with proline as the sole nitrogen source, and in late-exponential phase, a nitrogen pulse is given to activate TORC1), using luminescence as a readout [1]. To achieve this, each yeast strain to be evaluated was transformed with a reporter construct containing the firefly luciferase gene (Luc) replacing its endogenous RPL26A open reading frame (ORF); RPL26A was selected because it is known to be expressed upon TORC1 activation and does not cause growth defects upon deletion [1]. This original method allowed us to interrogate the genetic basis associated with TORC1 activation by phenotyping a recombinant biparental population composed of 96 segregant yeast strains and performing QTL mapping [15]. Nevertheless, this methodology had room for improvement to be used with larger yeast populations with greater genotypic diversity, such as those typically used for GWAS. First, each yeast strain transformed with the reporter construct had to be verified by polymerase chain reaction (PCR) to ensure that the reporter construct was in the correct genomic locus, which was time-consuming. Even more important, each strain had the Luc gene controlled by its endogenous RPL26A gene promoter (PRPL26A), so differences in luminescence could be due to differences in the PRPL26A sequence and not to differences in TORC1 activation.
The present protocol was developed on the study by Rocha et al. [11] as an improvement of the previous methodology [1], where a reporter plasmid, called the pTOMAN-G plasmid, was generated. The use of this plasmid also allows for indirectly monitoring the activation of the TORC1 signaling pathway through the expression of the Luc gene under the control of PRPL26A, but it overcomes the aforementioned limitations. First, in the pTOMAN-G plasmid, the Luc gene is under the control of the PRPL26A obtained from the S. cerevisiae laboratory strain BY4741 (a haploid derived from strain S288c, which is the reference strain of the species), which ensures consistent regulation of the promoter in all transformed strains as each one has the same PRPL26A controlling Luc expression. Additionally, the use of a plasmid carrying an antibiotic resistance gene as a selectable marker facilitates the confirmation of transformant yeast strains, with no need for the PCR step. As a proof of concept, this improved method allowed monitoring the activation of the TORC1 signaling pathway quickly and effectively in a large yeast population, derived from the 1002 Yeast Genomes Project (a project that fully sequenced 1011 yeast strains from different ecological niches) [16], giving rise to the TOMAN-G population (which is composed of 270 yeast strains transformed with the TOMAN-G plasmid) [11]. As presented here, this protocol is restricted to yeast strains capable of growth on proline as the sole nitrogen source (because nitrogen upshift experiments require it) but could be extended to other types of experiments (e.g., TORC1 signaling inhibition by rapamycin) that would allow for the assessment of other aspects of TORC1 activation in even more yeast strains. Although in principle the applicability and efficacy of this protocol are restricted to S. cerevisiae, because the plasmid design includes amplified elements from the BY4741 strain genome (such as the PRPL26A sequence), a redesign of the plasmid could extend its use as a reporter tool to monitor activation of the TORC1 pathway, or other pathways, in other yeast species.
Materials and reagents
Biological materials
1. Yeast strains to be evaluated
2. Saccharomyces cerevisiae BY4741 strain, genotype: MATa; his3∆1; leu2∆0; met15∆0; ura3∆0 (this yeast strain is available at ATCC®, catalog number: 201388)
3. Escherichia coli DH5α strain, competent cells, genotype: F– ϕ80lacZΔ M15 Δ (lacZYA-argF) U169 recA1 endA1 hsdR17 (rK– mK+) phoA supE44 λ- thi–1 gyrA96 relA1 (Thermo Scientific, catalog number: EC0112)
4. pRS316 plasmid [17] (available at ATCC®, catalog number: 77145)
5. pRS426 plasmid including the luciferase reporter (Luc), CYC1 terminator (CYCTER), and the hygromycin cassette (HphMx) [18]
Note: The pTOMAN-G plasmid is available upon request from the authors.
Reagents
1. BD DifcoTM yeast nitrogen base without amino acids and ammonium sulfate (BD Biosciences, catalog number: 233520)
2. Yeast nitrogen base without amino acids (BD Biosciences, catalog number: 291940)
3. BD DifcoTM LB broth, Lenox (BD Biosciences, catalog number: 240230)
4. Dropout medium without uracil, histidine, leucine, and tryptophan (USBiological, catalog number: D9540)
5. Yeast extract powder (HuanKai Microbial, catalog number: HAM009)
6. D-glucose, anhydrous (ChemCruz, catalog number: sc-211203A)
7. BactoTM peptone (GibcoTM, catalog number: 211677)
8. Agar-agar (MilliporeSigma, catalog number: 1016141000)
9. D-luciferin, potassium salt (GoldBio, catalog number: LUCK-1G)
10. L-proline (Sigma-Aldrich, catalog number: P5607-100G)
11. L-glutamine (Sigma-Aldrich, catalog number: G3126-100G)
Note: See General note 2.
12. L-leucine (Sigma-Aldrich, catalog number: L8000-25G)
13. L-histidine (Sigma-Aldrich, catalog number: H8000-5G)
14. L-tryptophan (Sigma-Aldrich, catalog number: T0254-5G)
15. Hygromycin B (50 mg/mL) (GibcoTM, catalog number: 10687010)
16. Enzyme phusion flash high-fidelity master mix (Thermo ScientificTM, catalog number: F548S); contains all the necessary reaction components for PCR, except for template DNA and primers
17. Sorbitol (Millipore, catalog number: 56755-1KG)
18. Glycerol (Santa Cruz Biotechnology, Inc., catalog number: sc-29095)
19. Sheared salmon sperm DNA (10 mg/mL) (InvitrogenTM, catalog number: AM9680)
20. Lithium acetate dihydrate (Sigma-Aldrich, catalog number: L6883-250G)
21. Poly(ethylene glycol) 3350 (PEG 3350) (Sigma-Aldrich, catalog number: 202444-250G)
22. GeneJET Plasmid Miniprep kit (Thermo ScientificTM, catalog number: K0503)
23. Ampicillin sodium salt (USBiological, catalog number: A2260)
24. BamHI restriction enzyme (New England BioLabs, catalog number: R0136S)
25. XhoI restriction enzyme (New England BioLabs, catalog number: R0146S)
26. Zymoprep Yeast Plasmid Miniprep I kit (Zymo Research, catalog number: D2001)
27. Wizard® Genomic DNA Purification kit (Promega, catalog number: A1120)
28. GoTaq green master mix (Promega, catalog number: M7122)
29. UltraPureTM DNase/RNase-free distilled water (Thermo ScientificTM, catalog number 10977-023)
Note: Referred to as UPdH2O in the protocol, to distinguish it from regular distilled water (dH2O)
30. Lafken agarose (Fermelo Biotec, catalog number: FER00AL200G)
31. TAE (Tris, acetic acid, EDTA) buffer (Winkler, catalog number: BM-0490)
32. Zymolyase 20T (MP Biomedicals, catalog number: 320921)
Solutions
1. L-proline stock solution (50×) (see Recipes)
2. L-glutamine stock solution (30×) (see Recipes)
3. D-luciferin stock solution (25×) (see Recipes)
4. Yeast minimal medium (YMM) (see Recipes)
5. YMM+Pro/YMM+Pro+Hyg/YMM+Pro+Luc+Hyg (see Recipes)
6. YPD medium/YPD-Agar+Hyg medium (see Recipes)
7. Sorbitol 1 M (see Recipes)
8. Glycerol 30% (v/v) (see Recipes)
9. Lithium acetate 1 M (see Recipes)
10. Lithium acetate 100 mM (see Recipes)
11. Salmon sperm DNA carrier 2.0 mg/mL (see Recipes)
12. PEG 50% (w/v) (see Recipes)
13. Synthetic complete (SC) medium minus uracil (see Recipes)
14. Zymolyase 10 mg/mL (see Recipes)
15. Lysogeny broth (LB) medium (see Recipes)
Recipes
1. L-proline stock solution (50×)
Mix 250 mg of L-proline in 10 mL of dH2O. Then, sterilize with a 0.22 μm syringe filter. The stock solution should be prepared up to one week in advance and stored at 4 °C until use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| L-proline | 25 mg/mL | 250 mg |
| dH2O | n/a | 10 mL |
2. L-glutamine stock solution (30×)
Mix 150 mg of L-glutamine in 10 mL of dH2O. Then, sterilize with a 0.22 μm syringe filter. The stock solution should be prepared up to one day in advance and stored at 4 °C until use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| L-glutamine | 15 mg/mL | 150 mg |
| dH2O | n/a | 10 mL |
Note: See General note 2.
3. D-luciferin stock solution (25×)
Mix 400 mg of D-luciferin in 50 mL of dH2O. Then, sterilize with a 0.22 μm syringe filter. Aliquot into 1.5 mL tubes (1 mL each), protect from light, and store at -20 °C. D-luciferin stock solutions are stable for at least 5 freeze-thaw cycles.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| D-luciferin | 25 mM | 400 mg |
| dH2O | n/a | 50 mL |
4. Yeast minimal medium (YMM)
Prepare 1 L of YMM. Mix 20 g of glucose and 1.71 g of yeast nitrogen base w/o amino acids and ammonium sulfate in 500 mL of dH2O. Then, complete to 1 L with dH2O and sterilize in an autoclave at 121 °C and 15 PSI for 15 min. YMM can be stored at room temperature for several months.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| D-glucose | 2% | 20 g |
| Yeast nitrogen base w/o amino acids and ammonium sulfate | 0.171% | 1.71 g |
| dH2O | n/a | 1 L |
5. YMM+Pro/YMM+Pro+Hyg/YMM+Pro+Luc+Hyg
Prepare 50 mL. Mix the components according to the solution to be used. Prepare for immediate use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| L-proline stock solution (Recipe 1) | 0.5 mg/mL | 1 mL |
| D-luciferin stock solution (Recipe 3) | 1 mM | 2 mL |
| Hygromycin B (50 mg/mL) | 0.3 mg/mL | 0.3 mL |
| YMM (Recipe 4) | n/a | To 50 mL |
6. YPD medium/YPD-Agar+Hyg medium
Prepare 500 mL of YPD medium. Mix the reagents: 10 g of glucose, 5 g of yeast extract, 10 g of peptone, and 10 g of agar-agar (optional) in 400 mL of dH2O. Then, complete to 500 mL with dH2O and sterilize in an autoclave at 121 °C and 15 PSI for 15 min. Optional: When the medium has cooled down to approximately 50–55 °C, add 3 mL of hygromycin B, mix, and pour into plates. YPD medium can be stored at room temperature for several months, whereas YPD-Agar+Hyg medium plates should be stored at 4 °C for up to 1 month.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| D-glucose | 2% | 10 g |
| Yeast extract | 1% | 5 g |
| Peptone | 2% | 10 g |
| Agar-agar (optional for plates) | 2% | 10 g |
| Hygromycin B (50 mg/mL) (optional for plates) | 0.3 mg/mL | 3 mL |
| dH2O | n/a | 500 mL |
7. Sorbitol 1 M
Prepare 100 mL of sorbitol 1 M. Mix 18.22 g of sorbitol in 80 mL of dH2O. Then, complete to 100 mL with dH2O and sterilize the solution using a 0.22 μm filter. Store at room temperature. Shelf life: 2 years.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Sorbitol | 1 M | 18.22 g |
| dH2O | n/a | 100 mL |
8. Glycerol 30% (v/v)
Prepare 100 mL of glycerol 30% (v/v). For this, measure 30 mL of glycerol using a graduated cylinder, add 60 mL of dH2O, and mix thoroughly. Transfer the solution to a 100 mL graduated cylinder and complete to 100 mL with dH2O. Sterilize in an autoclave at 121 °C and 15 PSI for 15 min. Store at room temperature. Shelf life: 3 years.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Glycerol | 30% | 30 mL |
| dH2O | n/a | 100 mL |
9. Lithium acetate 1 M
Prepare 100 mL of lithium acetate 1 M. Mix 10.2 g of lithium acetate in 80 mL of dH2O. Stir until the lithium acetate is completely dissolved and complete to 100 mL with dH2O. Sterilize in an autoclave at 121 °C and 15 PSI for 15 min. Store at room temperature. Shelf life: 2 years.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Lithium acetate | 1 M | 10.2 g |
| dH2O | n/a | 100 mL |
10. Lithium acetate 100 mM
Prepare 100 mL of lithium acetate 100 mM. Mix 10 mL of lithium acetate and 90 mL of sterile dH2O. Store at room temperature until ready to use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Lithium acetate | 100 mM | 1.02 g |
| Sterile dH2O | n/a | 100 mL |
11. Salmon sperm DNA carrier 2.0 mg/mL
Prepare 1 mL of salmon sperm DNA carrier solution at 2.0 mg/mL from the sheared salmon sperm DNA (10 mg/mL) stock. Store at -20 °C, ensuring to avoid repeated freeze-thaw cycles. The solution remains stable for up to one year.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Salmon sperm DNA carrier (10 mg/mL) | 2.0 mg/mL | 200 μL |
| UPdH2O | n/a | 800 μL |
12. PEG 50% (w/v)
Prepare 100 mL of PEG 50% (w/v). In a volumetric flask, dissolve 50 g of PEG 3350 in 60 mL of dH2O, stirring with a magnetic stirrer until fully dissolved. Adjust the volume to exactly 100 mL with dH2O. Transfer the solution to an airtight container and sterilize in an autoclave at 121 °C and 15 PSI for 15 min. Store the solution tightly sealed, protected from light. Shelf life: 1 year stored at 4 °C.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| PEG 3350 | 50% | 50 g |
| dH2O | n/a | 100 mL |
13. Synthetic complete (SC) medium minus uracil
Prepare 500 mL of SC medium minus uracil by mixing the reagents in 400 mL of dH2O. Stir until the reagents are fully dissolved, then adjust the volume to 500 mL with dH2O. Sterilize the medium in an autoclave at 121 °C and 15 PSI for 15 min. Optionally, pour plates for solid medium if required. Protect from light and prepare it for immediate use.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| YNB | 0.68% | 3.4 g |
| D-glucose | 2% | 10 g |
| Dropout medium without uracil, histidine, leucine, and tryptophan | 0.2% | 1 g |
| L-Histidine | 0.002% | 10 mg |
| L-Leucine | 0.01% | 50 mg |
| L-Tryptophan | 0.002% | 10 mg |
| Agar-agar (optional for plates) | 2% | 10 g |
| dH2O | n/a | 500 mL |
14. Zymolyase 10 mg/mL
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Zymolyase 20T | 10 mg/mL | 10 mg |
| Molecular biology nuclease-free dH2O | n/a | 1 mL |
15. Lysogeny broth (LB) medium
Prepare 500 mL of LB medium by mixing the reagents in 400 mL of dH2O. Stir until the reagents are fully dissolved, then adjust the volume to 500 mL with dH2O. Sterilize the medium in an autoclave at 121 °C and 15 PSI for 15 min. Optionally, pour plates for solid medium if required; if needed, when the medium has cooled down to approximately 50–55 °C, add 500 μL of ampicillin, mix, and pour into plates. LB medium can be stored at room temperature for several months.
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DifcoTM LB broth | 2% | 10 g |
| Agar-agar (optional for plates) | 1.5% | 7.5 g |
| Ampicillin sodium salt (100 mg/mL) (optional for bacterial selection, add it after the sterilization process) | 100 µg/mL | 500 μL |
| dH2O | n/a | 500 mL |
Laboratory supplies
1. Syringe filters, MCE membrane, 30 mm, 0.22 μm, sterile (Jet Biofil, catalog number: FMC201030-ZX)
2. 96-well plate transparent bottom, sterile (SPL Life Sciences, catalog number: 30096)
3. 96-well plate white/optical bottom, non-treated surface, no lid, non-sterile (Thermo ScientificTM, catalog number: 265302)
4. Electroporation cuvettes, 2 mm (Genesee Scientific, catalog number: 40-101)
5. Conical tubes, 15 mL (Jet Biofil, catalog number: CFT011150-ZX)
6. Conical tubes, 50 mL (Jet Biofil, catalog number: CFT011500-ZX)
7. 14 mL culture tubes (FalconTM, catalog number: 352057)
8. Pipette tips, 10 μL (Tarsons, catalog number: 521000)
9. Pipette tips, 200 μL (Tarsons, catalog number: 521010-Y)
10. Pipette tips, 1,000 μL (Tarsons, catalog number: 521020-B)
11. Microcentrifuge tubes, 1.5 mL (Servicebio, catalog number EP-150-J)
12. Multichannel micropipette, 12 × 20 (GilsonTM, catalog number: F144071)
13. Multichannel micropipette, 12 × 200 μL (GilsonTM, catalog number: F144073)
14. Micropipette, 2 μL (GilsonTM, catalog number: F144054M)
15. Micropipette, 20 μL (GilsonTM, catalog number: F144056M)
16. Micropipette, 200 μL (GilsonTM, catalog number: F144058M)
17. Micropipette, 1,000 μL (GilsonTM, catalog number: F144059M)
18. Sterile cryogenic tubes, 2 mL (SSI Bio, catalog number: 2341-S0)
19. Petri dish 90 × 14 mm (Tarsons, catalog number: 460095)
20. 0.2 mL PCR tubes (Citotest, catalog number: 5460-0001)
22. Parafilm (Amcor, catalog number: PM-996)
22. Disposable gloves (Health Touch, catalog number: GEE1222)
23. Smear loop (Deltalab, catalog number: 302755)
24. Cell spreader (Deltalab, catalog number: 200500.1)
Equipment
1. SynergyTM HTX multi-mode microplate reader (BioTek Instruments, Inc., model: S1LFA)
Note: The use of this equipment cannot be replaced by that of the SunriseTM microplate reader.
2. SunriseTM absorbance reader with temperature control (Tecan Group Ltd., model: Sunrise)
Note: The use of this equipment may be replaced by that of the SynergyTM HTX microplate reader.
3. Electroporation system (Bio-Rad Laboratories, Inc., model: Gene Pulser XcellTM)
4. Microcentrifuge refrigerated (Gyrozen Co., Ltd., model: 1730R)
5. Centrifuge for conical tubes (Kubota Corporation, model: S300TR)
6. Centrifuge with a rotor A6-50P for 50 and 15 mL tubes (DLAB, model: DM0412)
7. Thermal cycler (Bio-Rad, model: T100)
8. Incubator at 30 °C (Faithful, model: DH4000BII)
9. Incubator at 37 °C (Breed Elos, model: B055F230)
10. Shaker incubator at 30 °C (LabTech, model: LSI-3016R)
11. Shaker incubator at 37 °C (LABWIT, model: Benchtop)
12. Laminar flow cabinet (BIOBASE, model: BBS-V800)
13. Thermal bath (LabTech, model: LWB-106D)
14. Heat block (DLAB, model: HB120-S)
15. Power supplies (C.B.S. Scientific, model: 300 Series)
16. Electrophoresis chamber (Fermelo Biotec, model: H2)
17. UV/Vis spectrophotometer (MicroDigital, model: Nabi)
Software and datasets
1. BioTek Gen5 software (BioTek Instruments, Inc., version 3.16.10)
2. Microsoft Office (Microsoft Corporation, version 16.91)
Note: Microsoft Office is a relevant software, because the data generated in the SynergyTM HTX microplate reader is extracted as Excel files from the BioTek Gen5 software.
3. Online platform Benchling for in silico design and other bioinformatics analyses (https://www.benchling.com/)
4. GraphPad Prism software (GraphPad Software, LLC., version 7.04)
5. Sanger sequencing service (Macrogen Inc., Republic of Korea)
6. Primer synthesis (Macrogen Inc., Republic of Korea)
Procedure
文章信息
稿件历史记录
提交日期: Feb 6, 2025
接收日期: May 20, 2025
在线发布日期: Jun 3, 2025
出版日期: Jun 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Gómez, M., Rocha, G., Ruiz, D., Martínez, C., Salinas, F. and Kessi-Pérez, E. I. (2025). High-Throughput Indirect Monitoring of TORC1 Activation Using the pTOMAN-G Plasmid in Yeast. Bio-protocol 15(12): e5356. DOI: 10.21769/BioProtoc.5356.
分类
微生物学 > 微生物遗传学 > 质粒
细胞生物学 > 细胞信号传导 > 胞内信号传导
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link