- EN - English
- CN - 中文
Optimized Midgut Tissue Dissociation of Mosquitoes and Sandflies for High-Quality Single-Cell RNA Sequencing
蚊虫和白蛉中肠组织的优化解离方法:用于高质量单细胞RNA测序
(§Technical contact) 发布: 2025年06月20日第15卷第12期 DOI: 10.21769/BioProtoc.5352 浏览次数: 2396
评审: Pilar Villacampa AlcubierreAnonymous reviewer(s)

相关实验方案
髓系肿瘤中用于生殖系 DNA 分离的皮肤成纤维细胞标准化培养:一种从床旁到实验室的实用方法
Parampreet Kour [...] Pulkit Rastogi
2025年10月05日 1411 阅读
Abstract
Single-cell RNA sequencing has revolutionized molecular cell biology by enabling the identification of unique transcription profiles and cell transcription states within the same tissue. However, tissue dissociation presents a challenge for non-model organisms, as commercial kits are often incompatible, and current protocols rely on tissue enzymatic digestion for extended periods. Tissue digestion can alter cell transcription in response to temperature and the stress caused by enzymatic treatment. Here, we propose a protocol to stabilize RNA using a deep eutectic solvent (Vivophix, Rapid Labs) prior to tissue dissociation, thereby avoiding transcription changes induced by the process and preventing RNase activity during incubation. We validated this methodology for three medically important insect vectors: Anopheles gambiae, Aedes aegypti, and Lutzomyia longipalpis. Single-cell RNA sequencing using our insect midgut dissociation protocol yielded high-quality sequencing results, with a high number of cells recovered, a low percentage of mitochondrial reads, and a low percentage of ambient RNA—two hallmark standards of cell quality.
Key features
• This protocol stabilizes tissue RNA before dissociation, avoiding RNase-mediated RNA degradation and transcription changes during the dissociation process.
• Current protocols for insect midgut dissociation use enzymatic digestion of live tissues at temperatures that are incompatible with insect physiology.
• We validated this new stabilization-dissociation methodology for insect midgut, yielding high-quality single-cell RNA sequencing with high gene counts, low mitochondrial RNA, and minimal ambient RNA contamination.
Keywords: Tissue dissociation (组织解离)Graphical overview
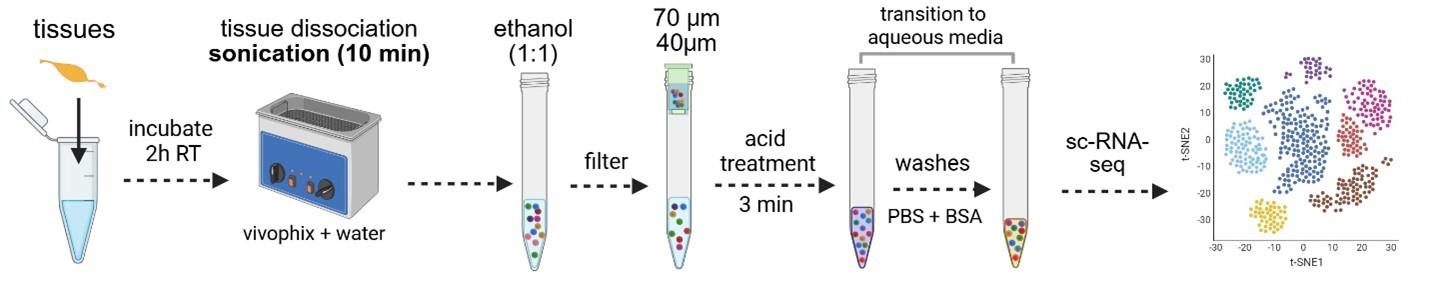
Mosquito and sand fly midgut RNA stabilization and dissociation protocol for single-cell sequencing applications
Background
Single-cell multi-omics approaches have become indispensable tools since they overcome the limitations of bulk sequencing to resolve cellular heterogeneity [1]. One advantage of these technologies is the ability to identify unique transcription profiles and different cell states within a single tissue; this requires cells to be dissociated and transformed into a single-cell suspension. However, dissociating cells while preserving nucleic acids, especially RNA, poses a significant challenge. RNA degradation can substantially affect downstream analysis, particularly in single-cell sequencing approaches that have less sequencing depth. While multiple dissociation methods that supposedly preserve RNA integrity before sequencing are available for mouse and human tissues, the same is not true for non-model organisms such as insects.
Insects are typically reared at 28 °C, a temperature within their physiological range. Still, the available methods for insect tissue dissociation, including those developed for Drosophila, involve enzymatic digestion of live tissues at room temperature [2–4] or high temperatures (37 °C) [5]. Therefore, in addition to the stress of enzymatic digestion, tissues are subjected to heat shock, which can lead to drastic transcriptional changes. Some methods describe the use of enzymes that work at low temperatures (4 °C) [6,7], but they require longer incubation periods, which can affect tissue oxygenation and viability, and consequently lead to transcriptional changes during sample processing. Furthermore, mosquito midguts express significant levels of RNases; therefore, longer incubation periods may also lead to RNA degradation. Of note, tissue dissociation can generate ambient RNA, which consists of cell-free RNA molecules present in the cell suspension, derived from ruptured, dead, or dying cells, and other sources [8,9]. Enzymatic digestion combined with mechanical disruption increases the amount of ambient RNA contamination in the sample, leading to a low fraction of quality reads in single cells [10].
Here, we propose an optimized protocol that dehydrates tissues and stabilizes RNA within cells before and during dissociation, thereby avoiding transcriptional changes caused by sample processing. Once the RNA is stabilized, dehydrated tissues can either be processed immediately or stored for up to several months. This is crucial for projects in which samples are harvested over time but need to be processed and sequenced simultaneously to avoid batch effects. It is also beneficial when the necessary infrastructure is not available or during field sample collection, because samples can be stored at room temperature, and the RNA remains stable.
The uniqueness and strength of this method rely on three key steps: 1) The dehydration of the tissue and stabilization of RNA within the cells using a deep eutectic solvent (presumably organic). This prevents any enzymatic reactions from occurring, keeping tissues stable and preserving their transcriptomes, although the tissues are no longer viable. After dissociation, the solvent is removed using ethanol. 2) Before transitioning to an aqueous medium, cells are incubated with acetic acid to inactivate any remaining RNase activity. 3) Dissociated cells are then washed in an aqueous medium and can be used for downstream single-cell RNA sequencing.
Using this procedure, we successfully dissociated midgut cells from three medically important insect vectors, preserving their RNA integrity and transcription patterns while achieving low ambient RNA contamination.
Materials and reagents
Biological materials
1. Anopheles gambiae (G3 – CDC strain) or Aedes aegypti (Liverpool strain) dissected midguts (30 midguts)
2. Lutzomya longipalpis (Jacobina strain) dissected midguts (50 midguts)
Reagents
1. Vivophix (RNA stabilization reagent for single cell analysis) (RapidLabs, UK, catalog number: RD-VIVO-50)
2. Nuclease-free water (NFW), no DEPC (Thermo Fisher Scientific, catalog number: AM2618)
3. RNASE inhibitor, protector RNASE inhibitor 10,000 units (Sigma-Aldrich, catalog number: 3335402001)
4. Phosphate buffer saline (PBS) 10× (90 g NaCl, 1.44 g KH2PO4, 7.95 g NaHPO4 in 1 L of water), pH 7.4. (KD-Medical, catalog number: RGF-3210)
5. Glacial acetic acid (Sigma-Aldrich, catalog number: A6283, 100 mL)
Caution: Flammable, skin burns/corrosion, eye damage, corrosive to metals. Handle inside of a safety cabinet.
6. Ethyl alcohol (Sigma-Aldrich, catalog number: E7023, 500 mL)
Caution: Flammable, irritant to skin and eyes.
7. Ultrapure BSA 50 mg/mL (Thermo Fisher Scientific, catalog number: AM2618)
8. RNase away (Molecular Bioproducts, catalog number: 7000)
9. 7-AAD (7-Aminoactinomycin D) (Thermo Fisher Scientific, catalog number: A1310) or Sytox (Thermo Fisher Scientific, catalog number: S7020)
10. Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: 472301)
11. WGA (wheat germ agglutinin) (Thermo Fisher Scientific, catalog number: W11261)
Solutions
1. Dissociation mixture (see Recipes)
2. Washing buffer (see Recipes)
3. Resuspension buffer (see Recipes)
4. Acid solution (see Recipes)
Recipes
Note: All recipes should be RNASE-free to keep RNA integrity during the procedure.
1. Dissociation mixture (per tube or sample)
| Reagent | Final proportion | Quantity or volume |
|---|---|---|
| Vivophix | 65% of final volume | 150 μL |
| Nuclease-free water | 35% of final volume | 350 μL |
Notes:
1. Add dissociation solution on top of 500 μL of Vivophix containing the sample to achieve the final proportion of 65/35).
2. Dissociation solution can be prepared proportionally to the number of samples.
3. Dissociation mixture cannot be stored and has to be prepared fresh before dissociation.
2. Washing buffer
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| PBS | 1× (150 mM, 1 mM KH2PO4, 5.5 mM NaHPO4) | 5 mL |
| Ultrapure BSA 50 mg/mL | 1 mg/mL or 0.1% | 1 mL |
| RNASE inhibitor (40 U/μL) | 1 U/50 mL | 1 μL |
| Nuclease-free water | n/a | 44 mL |
| Total | n/a | 50 mL |
Notes:
1. Each sample will require the use of 20 mL of washing buffer.
2. Because of the BSA and RNASE inhibitor, the washing buffer must be freshly prepared before dissociation. Storage is not recommended.
3. Resuspension buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PBS | 1× (150 mM, 1 mM KH2PO4, 5.5 mM NaHPO4) | 100 μL |
| Ultrapure BSA 50 mg/mL | 1 mg/mL or 0.1% | 20 μL |
| RNASE inhibitor (40 U/μL) | 0.2 U/μL | 5 μL |
| Nuclease-free water | n/a | 875 μL |
| Total | n/a | 1 mL |
Notes:
1. Each sample will require up to 200 μL.
2. Because of the BSA and RNASE inhibitor, the resuspension buffer must be freshly prepared before dissociation. Storage is not recommended.
4. Acid solution (per sample)
| Reagent | Final proportion | Quantity or volume |
|---|---|---|
| Vivophix | 60% | 600 μL |
| Glacial acetic acid | 40% | 400 μL |
| Total | n/a | 1 mL |
Notes:
1. The acid solution should preferably be freshly prepared and well mixed. However, it can be stored at room temperature for 72 h.
2. Acid solution can be prepared proportionally to the number of samples.
Caution: Prepare and handle this solution inside a safety cabinet.
Laboratory supplies
1. CellPro 15 mL conical centrifuge tubes (Alkali Scientific, catalog number: CW5600)
2. CellPro 50 mL conical centrifuge tubes (Alkali Scientific, catalog number: CW5603)
3. Open-top thick wall polycarbonate tubes, 8 × 51 mm, 25 Pk (1 mL) (Beckman Coulter, catalog number: 355657)
4. 1.5 mL microtubes (Axygen, catalog number: MCT-150-C-S)
5. 200 μL micropipette tips (TipOne 200 μL graduated filter tips) (USA Scientific, catalog number: 1180-8810)
6. 1250 μL micropipette tips (Purepoint barrier tips FT1250) (Alkali Scientific, catalog number: FT1250)
7. C-Chip disposable hemocytometer (iNCYTO, catalog number: DHC-N01-5 NI – Neubauer Improved)
9. 70 μm filter (Pluriselect, catalog number: 43-10040-70) or pre-separation filters (70 μm) (Miltenyl Biotec, catalog number: 130-095-823)
10. 40 μm filter (Pluriselect, catalog number: 43-10040-40)
11. MACS® SmartStrainers (30 μm) (Miltenyl Biotec, catalog number: 130-098-458)
12. Transfer pipette, sterile, polyethylene (Millipore Sigma, catalog number: Z350818-500EA)
13. Parafilm (to seal thick wall polycarbonate tubes) (Amcor, catalog number: PM-996)
14. Cellometer cell counting chamber (Nexcelom Bioscience, catalog number: SD100)
15. 15 μ-slide 18 well (Ibidi, catalog number: 81816) or any chamber that is compatible with a fluorescence microscope
16. Laboratory pipettes (10, 20, 200, and 1,000 μL)
Equipment
1. Professional benchtop ultrasonic cleaner, E-series, dual ultrasonic frequency 28/40 kHz
2. Thermo Sorvall Legend XTR refrigerated centrifuge (Thermo Fisher Scientific, catalog number: 75004520, discontinued)
3. TX-1000 4 × 1,000 mL swinging bucket rotor (Thermo Fisher Scientific, catalog number: 75003001)
Note: In this protocol, it is critical to use a swing bucket rotor type.
4. Cellometer Auto 2000 automated cell counter cell profiler (Nexcelom Bioscience, discontinued)
5. Transmitted light and epifluorescence microscopes
Procedure
文章信息
稿件历史记录
提交日期: Mar 10, 2025
接收日期: May 9, 2025
在线发布日期: Jun 3, 2025
出版日期: Jun 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Barletta, A. B. F., Talyuli, O. A., Cecilio, P. and Barillas-Mury, C. (2025). Optimized Midgut Tissue Dissociation of Mosquitoes and Sandflies for High-Quality Single-Cell RNA Sequencing. Bio-protocol 15(12): e5352. DOI: 10.21769/BioProtoc.5352.
分类
细胞生物学 > 组织分析 > 组织分离
细胞生物学 > 单细胞分析
系统生物学 > 转录组学 > RNA测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link










