- EN - English
- CN - 中文
Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Ub-POD:用于识别人细胞中E3泛素连接酶底物的泛素特异性邻近标记技术
(*contributed equally to this work) 发布: 2025年06月20日第15卷第12期 DOI: 10.21769/BioProtoc.5351 浏览次数: 2425
评审: Sébastien GillotinAmit Kumar DeyAnonymous reviewer(s)

相关实验方案
利用近端连接分析在新鲜分离的大鼠血管平滑肌细胞中定量Kv7.4与Dynein蛋白的相互作用
Jennifer van der Horst and Thomas A. Jepps
2024年03月20日 1562 阅读
Abstract
Ubiquitination is a post-translational protein modification that regulates a vast majority of processes during protein homeostasis. The covalent attachment of ubiquitin is a highly regulated process carried out by the sequential action of the three enzymes E1, E2, and E3. E3 ligases share a dual function of 1) transferring covalently attached ubiquitin from the catalytic cysteine of E2 (E2~Ub) to the substrate and 2) providing substrate specificity. Our current knowledge of their individual substrate pools is incomplete due to the difficult capture of these transient substrate–E3 ligase interactions. Here, we present an efficient protocol that enables the selective biotinylation of substrates of a given ubiquitin ligase. In brief, the candidate E3 ligase is fused to the biotin ligase BirA and ubiquitin to a biotin acceptor peptide, an Avi-tag variant (-2) AP. Cells are co-transfected with these fusion constructs and exposed to biotin, resulting in a BirA-E3 ligase-catalyzed biotinylation of (-2) AP-Ub when in complex with E2. As the next step, the biotinylated (-2) AP-Ub is transferred covalently to the substrate lysine, which enables an enrichment via denaturing streptavidin pulldown. Substrate candidates can then be identified via mass spectrometry (MS). Our ubiquitin-specific proximity-dependent labeling (Ub-POD) method allows robust biotinylation of the ubiquitylation substrates of a candidate E3 ligase thanks to the wild-type BirA and biotin acceptor peptide fused to the E3 and Ub, respectively. Because of the highly Ub-specific labeling, Ub-POD is more appropriate for identifying ubiquitination substrates compared to other conventional proximity labeling or immunoprecipitation (IP) approaches.
Key features
• A simple and cost-effective method using common chemicals makes Ub-POD easy to implement in any laboratory.
• Can be an exploratory tool to identify new substrates via mass spectrometry (MS) or as a validation tool in combination with immunoblotting or immunofluorescence.
• Knowledge of triggers and constraints of E3 ligase activity is beneficial.
Keywords: Ub-POD (Ub-POD)Graphical overview
Background
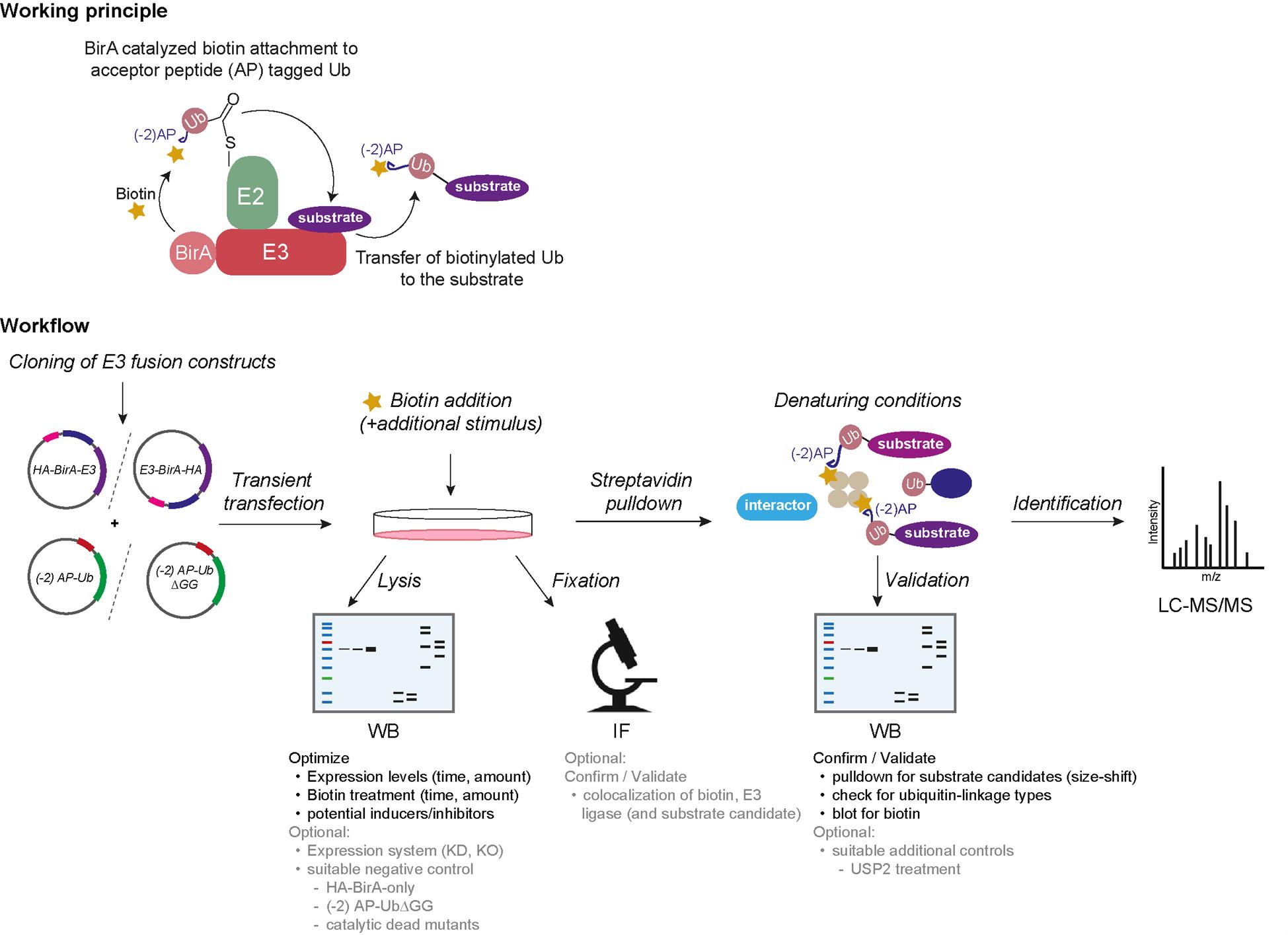
Ubiquitination is a post-translational protein modification with a pivotal role in many cellular processes, including the regulation of protein stability, enzymatic activity, cellular protein localization, and transport. Altogether, it leads to a functional conversion of proteins and regulates signaling events in a time- and spatial-dependent manner. Ubiquitin is a 76 amino acid small protein conjugated to its target by a concerted three-step enzymatic cascade, comprising the E1 activating enzyme, the E2 conjugating enzyme, and the E3 ligase [1]. The reaction is initiated with the activation of ubiquitin by one of the two E1 enzymes, which adenylates the terminal carboxyl group of ubiquitin prior to formation of a high-energy thioester bond with the catalytic cysteine of E1. Ubiquitin is then passed on to one of ~40 members of E2 via an E2-catalyzed transthioesterification reaction. Lastly, the E3 ligase interacts with the E2~Ub complex as well as with its substrate and enables the covalent attachment of the terminal carboxyl group of ubiquitin to its substrate lysine through an isopeptide bond [1–3]. Over 600 different E3 ligases are estimated to convey substrate specificity and make ubiquitination a very versatile system with over thousands of targets. E3 ligases can furthermore catalyze ubiquitination of ubiquitin itself at several lysine residues (K6, K11, K48, K63, etc.), resulting in different ubiquitin linkage chain types. The linkage type can often be interpreted as a code for different protein fates. For instance, K48 ubiquitin linkage chain types are often utilized as signals for proteasomal degradation, whereas K63 chains tend to have rather regulatory functions [3]. To understand the cellular function of an E3 ligase, it is thus inevitable to characterize its substrates and their respective regulation upon ubiquitination. However, due to the extremely transient nature of the E3 ligase–substrate interaction, conventional interaction-based methods [e.g., immunoprecipitation (IP), yeast two-hybrid] often fail to capture E3 ligase substrate pools completely [4]. On the other hand, powerful techniques such as diGly remnant profiling are elaborate and expensive [4,5]. To tackle this issue, we developed a simple and cost-effective method, which exploits the close proximity of the candidate E3 ligase to both its substrate and the E2~Ub complex. We use a proximity-based labeling approach, which is dependent on ubiquitin (Ub-POD) by tweaking the BioID principle with two essential changes [6]. First, we tagged our candidate E3 ligase with wildtype E. coli biotin ligase BirA instead of its highly reactive but promiscuous mutant BirA* (R118G mutation) to restrain its activity to BirA’s acceptor peptide (AP) substrate only. Second, we attached a modified acceptor peptide (-2) AP of BirA to the N-terminus of ubiquitin, rendering it to be selectively biotinylated by BirA WT in a proximity-dependent manner. We then co-transfected these constructs in cells and exposed them to biotin. Upon interaction of the E3 ligase with the E2~ (-2) AP-Ub complex as well as with its substrate, (-2) AP ubiquitin gets biotinylated and transferred to its target, leading to biotin-labeled ubiquitinated substrates. These can be pulled down from cell lysates under denaturing conditions, followed by either digestion of proteins for identification via mass spectrometry (MS) or immunoblotting for substrate validation.
Materials and reagents
Biological materials
1. Target cell line, e.g., human embryonic kidney epithelial cell line 293 (HEK-293) (American Type Culture Collection CRL-1573) (see General note 4)
Reagents
1. Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, catalog number: 41965039)
2. Trypsin-EDTA (0.25%), phenol red (Gibco, catalog number: 25200072)
3. Fetal bovine serum (FBS) (Gibco, catalog number: 10270106)
4. Phosphate buffered saline (PBS) (1×) (Gibco, catalog number: 14190169)
5. Opti-MEM (Gibco, catalog number: 31985062)
6. PCR reagents:
a. 5× Phusion HF buffer (New England Biolabs, catalog number: B0518)
b. Phusion HF DNA polymerase (New England Biolabs, catalog number: M0530)
c. dNTP solution mix (New England Biolabs, catalog number: N0447)
d. DMSO (New England Biolabs, catalog number: B0515)
e. Nuclease-free water (New England Biolabs, catalog number: B1500)
7. Template plasmids/cDNAs of the candidate E3 ligases
8. BirA vectors [Addgene, catalog numbers: 232584 (Empty BirA), 232586 (for tagging BirA at the N terminus of the ligase; there is a GSGS linker between BirA and the ligase), 232587 (for tagging BirA at the C terminus of the ligase), and 232588 (for tagging BirA at the C terminus of the ligase; there is a GSGS linker between BirA and the ligase)] and Avi-tagged Ub constructs [Addgene, catalog numbers: 232577, 232582 (this modified Avi-tagged Ub construct (-2)AP-Ub has been used to describe the protocol and also has been used in the original publication), and 232583] (plasmids can be obtained from the following source: https://www.addgene.org/Sagar_Bhogaraju/)
9. Transfection reagents, e.g., polyethyleneimine (PEI) (Polysciences, catalog number: 23966-1), Lipofectamine 2000 (Invitrogen, catalog number: 11668027), or Lipofectamine 3000 (Invitrogen, catalog number: L3000008)
Note: For optimal results, the transfection reagent should be selected based on the transfection efficiency of the specific cell line of interest.
10. Biotin (Sigma-Aldrich, catalog number: B4639)
11. MG132 (Calbiochem, catalog number: 474787)
12. Streptavidin agarose (Pierce/Thermo Scientific, catalog number: 20353)
13. Albumin fraction V (BSA) (Carl Roth, catalog number: 8076.3)
14. Benzonase® nuclease (Millipore, catalog number: E1014)
15. cOmpleteTM, mini protease inhibitor cocktail (Roche, catalog number: 11836153001)
16. Dithiothreitol (DTT) (MP Biomedical, catalog number: 856126)
17. N-ethylmaleimide (NEM) (Pierce/Thermo Scientific, catalog number: 23030)
18. Tris base [tris(hydroxymethyl)aminomethane] (Euromedex, catalog number: 200923A)
19. Glycine (Carl Roth, catalog number: 56-40-6)
20. Sodium chloride (NaCl) (Euromedex, catalog number: 1112-A)
21. Triton X-100 (Thermo Scientific Alfa Aesar, catalog number: A16046.AE)
22. SDS (Serva, catalog number: 20765)
23. EGTA (Millipore, catalog number: 324626)
24. Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma, catalog number: M2670)
25. Tween-20 (Carl Roth, catalog number: 9127.1)
26. 4× Laemmli sample buffer (Bio-Rad, catalog number: 1610747)
27. 2× Laemmli sample buffer (Bio-Rad, catalog number: 161-0737)
Note: 2-mercaptoethanol should be added freshly before use to a final concentration of 355 mM to both Laemmli sample buffers.
28. Color pre-stained protein standard, broad range (10–250 kDa) (NEB, catalog number: P7719)
29. Immobilon Crescendo Western HRP substrate (Millipore, catalog number: WBLUR)
30. Poly-D-Lysine (Gibco, catalog number: A3890401)
31. Paraformaldehyde solution, 4% in PBS (Thermo Scientific, catalog number: J61899.AK)
32. ProLongTM diamond antifade mountant with DAPI (Invitrogen, catalog number: P36966)
33. PR-619 [Tocris (Biotechne), catalog number: 4482]
34. USP2 (R&D Systems, catalog number: E504)
35. 2-Mercaptoethanol (Gibco, catalog number: 21985023)
36. Sodium phosphate, dibasic (Na2HPO4·2H2O) (Carl Roth, catalog number: 4984.1)
37. Potassium phosphate, monobasic (KH2PO4) (Carl Roth, catalog number: 3904.1)
Antibodies
1. IRdye streptavidin 680 (Licor, catalog number: 926-32230)
2. IRdye streptavidin 800 (Licor, catalog number: 926-68079)
3. HA-Tag (C29F4) rabbit mAb (Cell Signaling Technology, catalog number: 3724-S)
4. Avi-tag (Genscript, catalog number: A00674)
5. Alpha-Tubulin (anti-mouse) (Bio-Techne Ltd., catalog number: NB100-690SS)
6. Goat anti-mouse IgG (H+L) secondary antibody, HRP (SantaCruz, catalog number: sc-516102)
7. Goat anti-rabbit IgG (H+L) secondary antibody, HRP (SantaCruz, catalog number: sc-2357)
Solutions
1. 1 M DTT (see Recipes)
2. 1 mM Biotin (see Recipes)
3. 10× phosphate buffered saline (PBS) (pH 7.4) (see Recipes)
4. 1 M Tris HCl, pH 7.5 (see Recipes)
5. 5 M NaCl (see Recipes)
6. 10% SDS (see Recipes)
7. 100 mM EGTA (see Recipes)
8. 1 M MgCl2·6H2O (see Recipes)
9. 2 M NEM (see Recipes)
10. 1× Cell Lysis Buffer (CLB) (see Recipes)
11. IP Wash buffer 1 (see Recipes)
12. IP Wash buffer 2 (see Recipes)
13. IP Wash buffer 3 (see Recipes)
14. 10× running buffer (see Recipes)
15. 10× transfer buffer (see Recipes)
16. 1× PBST (see Recipes)
17. Blocking buffer (see Recipes)
18. Antibody dilution buffer for streptavidin immunoblotting (see Recipes)
19. Blocking and permeabilization buffer for streptavidin immunofluorescence microscopy (see Recipes)
20. Antibody dilution buffer/wash buffer for streptavidin immunofluorescence microscopy (see Recipes)
Recipes
1. 1 M DTT
Dissolve 1.5425 g of DTT in 8 mL of distilled water. Adjust volume to 10 mL. Filter through a 0.22 μm filter. Aliquot and store in the dark at -20 °C.
2. 1 mM Biotin
Dissolve 12.2155 mg of biotin powder in 50 mL of serum-free DMEM. Vortex. Filter through a 0.22 μm filter. Make aliquots to prevent freeze-thawing.
3. 10× phosphate buffered saline (PBS) (pH 7.4)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 1.37 M | 80 g |
| KCl | 27 mM | 2 g |
| Na2HPO4⋅2H2O | 100 mM | 17.8 g |
| KH2PO4 | 18 mM | 2.4 g |
| Distilled water | n/a | Up to 1 L |
Dissolve all the reagents in 800 mL of distilled water. Adjust the pH to 7.4. Fill the volume up to 1 L with distilled water. Filter through a 0.22 μm filter and store at room temperature (RT) or 4 °C.
4. 1 M Tris HCl (pH 7.5)
Dissolve 121.14 g of Tris base in 800 mL of distilled water. Adjust to the desired pH with concentrated HCl. Mix and add distilled water to 1 L. Sterilize by autoclaving and store at RT.
5. 5 M NaCl
Dissolve 292.2 g of NaCl powder in 1 L of distilled water. Sterilize by autoclaving and store at RT.
6. 10% SDS
Dissolve 10 g of SDS in 80 mL of distilled water. Fill the volume up to 100 mL. Dissolve the solution on a magnetic stirrer. If needed, heat the solution slightly to dissolve the powder. Store at RT.
7. 100 mM EGTA
Dissolve 3.804 g of EGTA in 10 mL of 1.7 M Tris solution. Add 80 mL of distilled water and adjust pH to 7.0 with 1.7 M Tris solution. Bring the final volume to 100 mL with distilled water. The solution may be stored at RT after sterilizing by autoclaving.
8. 1 M MgCl2·6H2O
Dissolve 20.33 g of MgCl2⋅6H2O in 100 mL of distilled H2O. This solution is extremely hygroscopic. Do not store open bottles for longer periods. Sterilize by autoclaving and store at RT.
9. 2 M NEM
Dissolve 12.513 g of NEM in 50 mL of isopropanol. Aliquot and store at -20 °C.
10. 1× cell lysis buffer (CLB)
| Reagent | Final concentration | Volume |
|---|---|---|
| 1 M Tris-HCl, pH 7.5 | 50 mM | 2.5 mL |
| 5 M NaCl | 150 mM | 1.5 mL |
| Triton X-100 | 1% | 500 μL |
| 10% SDS | 0.1% | 500 μL |
| 100 mM EGTA | 1 mM | 500 μL |
| 1 M MgCl2⋅6H2O | 1.5 mM | 75 μL |
| Benzonase | 250 U/mL | 50 μL |
| 2 M NEM | 50 mM | 1.25 mL |
| Protease inhibitor cocktail | n/a | 1 tablet/50 mL |
| MilliQ water | n/a | up to 50 mL |
Store at -20 °C.
11. IP wash buffer 1
| Reagent | Final concentration | Volume |
|---|---|---|
| 1 M Tris-HCl, pH 7.5 | 50 mM | 2.5 mL |
| 5 M NaCl | 500 mM | 5 mL |
| Triton X-100 | 1% | 500 μL |
| 10% SDS | 1% | 5 mL |
| 1 M DTT | 10 mM | 500 μL |
| 100 mM EGTA | 1 mM | 500 μL |
| 2 M NEM | 50 mM | 1.25 mL |
| Protease inhibitor cocktail | n/a | 1 tablet/50 mL |
| MilliQ water | n/a | up to 50 mL |
Add SDS and DTT freshly before use.
12. IP wash buffer 2
| Reagent | Final concentration | Volume |
|---|---|---|
| 1 M Tris-HCl, pH 7.5 | 50 mM | 2.5 mL |
| 5 M NaCl | 150 mM | 1.5 mL |
| Triton X-100 | 1% | 500 μL |
| 10% SDS | 0.1% | 500 μL |
| 1 M DTT | 10 mM | 500 μL |
| 100 mM EGTA | 1 mM | 500 μL |
| 2 M NEM | 50 mM | 1.25 mL |
| Protease inhibitor cocktail | n/a | 1 tablet/50 mL |
| MilliQ water | n/a | up to 50 mL |
Add DTT freshly before use.
13. IP Wash buffer 3
| Reagent | Final concentration | Volume |
|---|---|---|
| 1 M Tris-HCl, pH 7.5 | 50 mM | 2.5 mL |
| 5 M NaCl | 150 mM | 1.5 mL |
| 2 M NEM | 50 mM | 1.25 mL |
| Protease inhibitor cocktail | n/a | 1 tablet/50 mL |
| MilliQ water | n/a | up to 50 mL |
14. 10× running buffer
| Reagent | Final concentration | Quantity |
|---|---|---|
| Tris base | 250 mM | 30.285 g |
| Glycine | 1.92 M | 144.4 g |
| SDS | 1% (w/v) | 10 g |
| Distilled water | n/a | up to 1 L |
Store at RT and dilute to 1× before use.
15. 10× transfer buffer
| Reagent | Final concentration | Quantity |
|---|---|---|
| Tris base | 250 mM | 30.285 g |
| Glycine | 1.92 M | 144.4 g |
| Distilled water | n/a | Up to 1 L |
Before use, make 1× transfer buffer by diluting 10× transfer buffer and adding 20% methanol.
16. 1× PBST
| Reagent | Final concentration | Volume |
|---|---|---|
| 10× PBS | 1× | 100 mL |
| Tween 20 | 0.2% | 2 mL |
| Distilled water | n/a | up to 1 L |
17. Blocking buffer
| Reagent | Final concentration | Quantity |
|---|---|---|
| BSA | 5% | 5 g |
| 1× PBS | n/a | 100 mL |
Store at 4 °C.
18. Antibody dilution buffer for streptavidin immunoblotting
| Reagent | Final concentration | Quantity |
|---|---|---|
| BSA | 5% | 0.5 g |
| 1× PBS | n/a | 10 mL |
| Tween-20 | 0.2% | 20 μL |
| 10 % SDS | 0.1% | 100 μL |
Add SDS and Tween-20 freshly before use.
19. Blocking and permeabilization buffer for streptavidin immunofluorescence microscopy
| Reagent | Final concentration | Quantity |
|---|---|---|
| BSA | 5% | 5 g |
| Triton X-100 | 0.3% | 300 μL |
| 1× PBS | n/a | up to 100 mL |
Store at 4 °C.
20. Antibody dilution buffer/wash buffer for streptavidin immunofluorescence microscopy
| Reagent | Final concentration | Quantity |
|---|---|---|
| BSA | 3% | 15 g |
| Triton X-100 | 0.1% | 500 μL |
| 1× PBS | n/a | up to 500 mL |
Store at 4 °C.
Laboratory supplies
1. 10 cm cell culture dishes (Corning, catalog number: 430167)
2. 6-well cell culture plates (Corning, catalog number: 353046)
3. Cell lifter/scraper (Biologix, catalog number: 70-2180)
4. Microtubes 0.5 mL (Treff, catalog number: 61-9708-73), 1.5 mL (Eppendorf, catalog number: 0030120086), and 2 mL (Eppendorf, catalog number: 0030120094)
5. 0.2 mL PCR tubes (StarLab, catalog number: I1402-8200)
6. Falcon centrifuge tubes, 15 mL (Corning, Falcon, catalog number: 352096) and 50 mL (Corning, Falcon, catalog number: 352070)
7. 4%–20% Mini-PROTEAN® TGX precast protein gels, 10-well (Bio-Rad, catalog number: 4561093)
8. Microscope slides (EprediaTM, catalog number: 17244884)
9. Cover glass (Fisherbrand, catalog number: 12333138)
Equipment
1. SDS-PAGE electrophoresis apparatus and chambers (Bio-Rad, catalog number: 1658004)
2. SDS-PAGE transfer apparatus (Bio-Rad, catalog number: 1703935)
3. Thermoblock (Thermo Fisher Scientific, catalog numbers: 13687717, 13687718)
4. Thermomixer (Boekel Scientific, catalog numbers: 270811, 270812)
5. Sonicator (Branson 450 Digital Sonifier; Bioruptor® Pico sonication device, catalog number: B01080010)
6. Cell culture incubator (HeracellTM 150i CO2 Incubator, 150 L, Thermo ScientificTM, catalog number: 51032720)
7. Gentle rotating mixer for Eppendorf tubes (end-over-end rotation) (VWR, catalog number: 10136-084)
8. Centrifuge for 15 and 50 mL tubes (Eppendorf, model: 5804)
9. Centrifuge for 1.5 mL reaction tubes with cooling function (Eppendorf, model: 5427 R)
10. Micro-pipettes (Gilson, catalog number: F167300)
11. NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000, catalog number: ND-2000)
12. PCR machine (Bio-Rad, model: T100 THERMAL CYCLER, catalog number: 1861096)
13. -80 °C freezer
14. -20 °C freezer
15. Refrigerator (4 °C)
16. Chemidoc MP imaging system (Bio-Rad, catalog number: 12003154)
17. Confocal microscope with 63× oil-immersion objective (Leica, model: TCS SP5)
Software and datasets
1. Image Lab software v5.2.1 (Bio-Rad)
2. Excel (Microsoft)
3. GraphPad Prism 9 (GraphPad)
4. ImageJ (Version 1.49s, National Institutes of Health)
5. Adobe Illustrator CS6 (Version 16.0.3)
Procedure
文章信息
稿件历史记录
提交日期: Jan 31, 2025
接收日期: May 11, 2025
在线发布日期: Jun 5, 2025
出版日期: Jun 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mukhopadhyay, U., Levantovsky, S., Behrends, C. and Bhogaraju, S. (2025). Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells. Bio-protocol 15(12): e5351. DOI: 10.21769/BioProtoc.5351.
- Mukhopadhyay, U., Levantovsky, S., Carusone, T. M., Gharbi, S., Stein, F., Behrends, C. and Bhogaraju, S. (2024). A ubiquitin-specific, proximity-based labeling approach for the identification of ubiquitin ligase substrates. Sci Adv. 10(32): eadp3000. https://doi.org/10.1126/sciadv.adp3000
分类
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
生物化学 > 蛋白质 > 降解
分子生物学 > 蛋白质 > 泛素化
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link