- EN - English
- CN - 中文
In vitro Condensation Assay of Fluorescent Protein-Fused PRPP Amidotransferase Purified from Budding Yeast Cells
源自出芽酵母的荧光蛋白融合PRPP转酰胺酶的体外冷凝实验
发布: 2025年06月05日第15卷第11期 DOI: 10.21769/BioProtoc.5335 浏览次数: 1774
评审: Abhilash PadavannilJoyce ChiuAnonymous reviewer(s)
Abstract
De novo synthesis of purine nucleotide is essential for the production of genetic materials and cellular chemical energy. PRPP amidotransferase (PPAT) is the rate-limiting enzyme in de novo purine synthesis, thereby playing a crucial regulatory role in this pathway. Recent studies suggest that metabolic enzymes, including PPAT, form condensates through phase separation to regulate cellular metabolism in response to environmental changes. However, due to the lack of methods for purifying eukaryotic PPAT, the biophysical properties of the enzyme have remained unknown. Here, I describe a protocol for purifying budding yeast PPAT tagged with green fluorescent protein from yeast cells, as well as an in vitro assay to examine condensation of the fluorescent PPAT by microscopy. These techniques enabled us to elucidate the mechanism controlling PPAT condensation and may also be applicable to the purification and condensation assay of other enzymes.
Key features
• First example of purification of fluorescent protein-fused PPAT.
• Time-saving, one-step affinity purification without expensive equipment.
• Automated image and data processing increases throughput and reproducibility and ensures reliability in the study of biomolecular condensates.
• A powerful alternative for protein purification to bacterial expression systems.
Keywords: Phase separation (相分离)Graphical overview
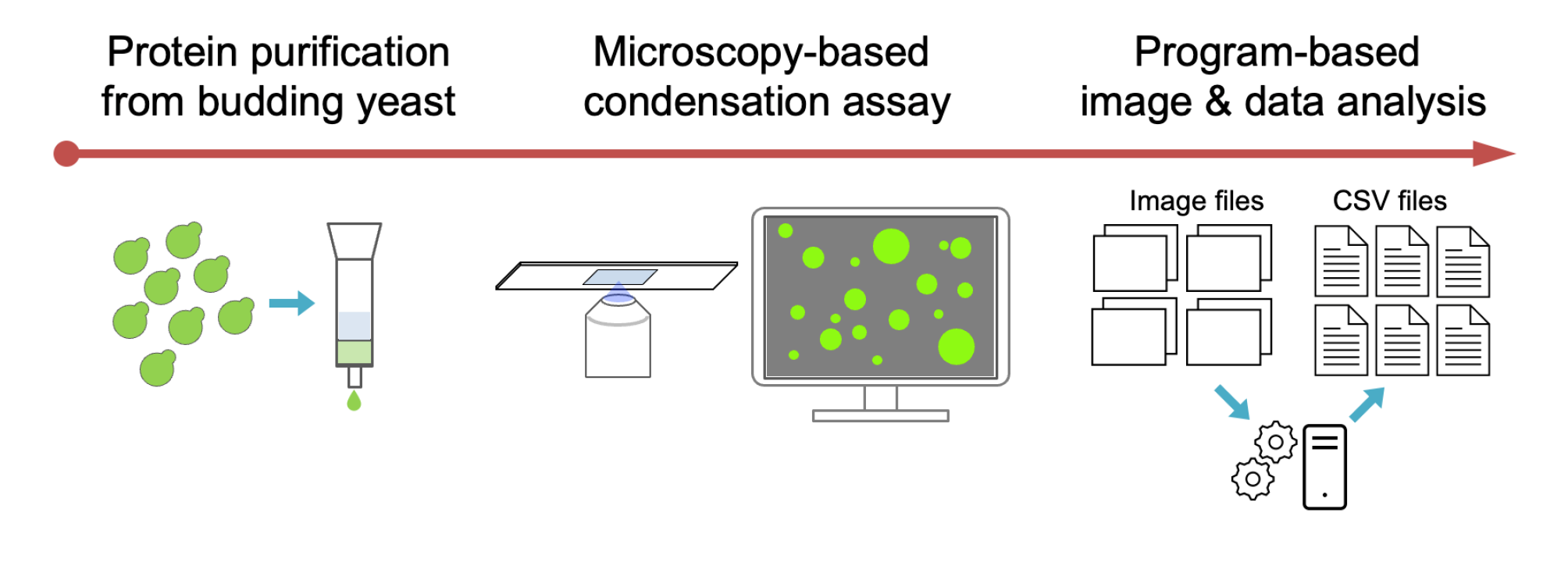
Workflow for purification of the fluorescent protein fused enzyme and in vitro condensation assay
Background
Purine nucleotides and their derivatives play crucial roles in various cellular functions, making them indispensable metabolites for all living organisms. These nucleotides are synthesized through both the salvage pathway and the de novo synthesis pathway. Under typical cellular conditions, the salvage pathway predominates, utilizing phosphoribosyl pyrophosphate (PRPP) to convert purine bases—often taken up from the extracellular environment—into purine nucleotides. However, when purine availability is limited or demand is elevated, such as in rapidly dividing cells, the de novo pathway is activated to ensure an adequate nucleotide supply [1]. Despite extensive research on the enzymes involved in purine biosynthesis, the precise regulatory mechanisms governing de novo purine synthesis remain incompletely understood.
PRPP amidotransferase (PPAT) (EC 2.4.2.14) catalyzes the initial and rate-limiting step of de novo purine biosynthesis, exerting a key regulatory function in this pathway. The enzyme’s biochemical properties and catalytic mechanisms have been extensively studied for decades, particularly through research using bacterial enzymes [2,3]. A notable characteristic of PPAT is its feedback inhibition by the end products of the de novo pathway, such as AMP [2–4]. This regulatory mechanism is thought to modulate de novo purine biosynthesis in response to extracellular purine availability, as observed in mammalian cell cultures [4]. However, the purification of eukaryotic PPAT has not yet been established, and its biochemical and biophysical properties remain largely uncharacterized.
A growing body of evidence suggests that many metabolic enzymes form condensates within cells in response to external stimuli to regulate cellular metabolism [5]. PPAT also reversibly localizes to cytoplasmic condensates upon depletion of extracellular purine bases in budding yeast and HeLa cells [6, 7]. However, attempts to express and purify yeast PPAT in bacteria have been unsuccessful [8], leaving the mechanism regulating yeast PPAT condensate formation and its physiological significance largely unknown.
Recently, we demonstrated that budding yeast PPAT, Ade4, self-assembles into micron-sized condensates through phase separation under molecular crowding conditions, leading to enzyme activation [9]. Purine nucleotides inhibit condensate formation, whereas PRPP promotes condensation, counteracting the inhibitory effects of purine nucleotides. To investigate this phenomenon, I developed a protocol to purify Ade4 tagged with the green fluorescent protein monomeric NeonGreen (mNG) from budding yeast cells using the Strep-tag®/Strep-Tactin® system [10], as well as a microscopy-based in vitro assay to examine the condensation of the fluorescent Ade4 protein. To our knowledge, this represents the first successful purification of a eukaryotic PPAT. This protocol can thus be adapted for the purification and condensation assays of PPAT from other species. Additionally, it offers a promising alternative for enzyme purification when bacterial expression proves unsuccessful, as was the case for yeast PPAT. This protocol is also useful when bacterial expression is inappropriate, for example, to investigate post-translational modifications of proteins specific to eukaryotic cells.
Several methods are available to study protein condensation in vitro, including bulk assays such as turbidity measurements and centrifugation, as well as fluorescence microscopy–based assays. Fluorescence microscopy–based assays, including the one introduced here, are especially useful as they allow direct observation of individual condensates, providing insights into condensate size, number, shape, and dynamic behavior. They can also reveal liquid-like properties and enable co-localization analysis by labeling different proteins with distinct fluorescent colors. Despite their strengths, fluorescence microscopy–based assays have limitations: they are low-throughput, less quantitative without image analysis, may miss small condensates, and often require labeling. Still, they remain key tools for understanding the mechanisms of protein condensation.
Materials and reagents
Biological materials
1. MTY3944 (MATa natNT2-PGPD-Ade4-mNG-Strep-tag II-hphNT1 his3∆1 leu2∆0 ura3∆0) (YGRC, BY29686)
2. MTY3969 (MATα his3Δ1::pRS303-PGPD-mNG-Strep-tag II-hphNT1 leu2Δ0 lys2Δ0 ura3Δ0) (YGRC, BY29688)
Note: MTY3944 and MTY3969 constitutively overexpress Ade4-mNG-Strep-tag II and mNG-Strep-tag II under the GPD promoter, respectively. Both yeast strains are available from the Yeast Genetic Resource Centre Japan (YGRC, http://yeast.nig.ac.jp/yeast/).
Reagents
1. Yeast extract (BD, catalog number: 212750)
2. Bacto-peptone (BD, catalog number: 211677)
3. D(+)-glucose (FUJIFILM Wako, catalog number: 049-31165)
4. Agar (FUJIFILM Wako, catalog number: 010-08725)
5. Adenine sulfate (FUJIFILM Wako, catalog number: 018-19613)
6. NaCl (FUJIFILM Wako, catalog number: 191-01665)
7. MgCl2·6H2O (FUJIFILM Wako, catalog number: 132-00175)
8. 0.5 M EDTA (DOJINDO, catalog number: 347-07481)
9. Ethanol, 99.5% (FUJIFILM Wako, catalog number: 057-00451)
10. 1 M Tris-HCl (pH 7.5) (NIPPON GENE, catalog number: 316-90221)
11. (+/-)-Dithiothreitol (FUJIFILM Wako, catalog number: 047-08973)
12. Benzylsulfonyl fluoride (aka phenylmethylsulfonyl fluoride, PMSF) (FUJIFILM Wako, catalog number: 028-15373)
13. PEG 8000 (FUJIFILM Wako, catalog number: 596-09755)
14. Buffer E (10×) (IBA GmbH, catalog number: 2-1000-025)
15. cOmplete protease inhibitor cocktail mini (EDTA-free) (Roche, catalog number: 11836170001)
16. Neutralized avidin (FUJIFILM Wako, catalog number: 015-24231)
17. Strep-Tactin Sepharose, 50% suspension (IBA GmbH, catalog number: 2-1201-002)
18. FastGene Q-stain (Nippon Genetics, catalog number: NE-FG-QS1)
19. Amicon Ultra-0.5 centrifugal filter device with a molecular weight cutoff of 30 kDa or 10 kDa [Merck, catalog number: UFC503024 (30 kDa) and UFC501008 (10kDa)]
20. Buffer R (10×) (IBA GmbH, catalog number: 2-1002-100)
21. Bovine serum albumin (BSA) standard solution (2 mg/mL) included in the Bradford Protein Assay Kit (TaKaRa, catalog number: T9310A)
Solutions
1. Adenine stock (see Recipes)
2. YPD agar plate (see Recipes)
3. YPDA (see Recipes)
4. 10× EB (see Recipes)
5. 0.5 M DTT (see Recipes)
6. EBD (see Recipes)
7. 0.1 M PMSF (see Recipes)
8. EBDPI (see Recipes)
9. EBDPIP (see Recipes)
10. 1 M MgCl2 (see Recipes)
11. EBMD (see Recipes)
12. EBM10D (see Recipes)
13. 50% PEG8000 (see Recipes)
14. 20% PEG8000 (see Recipes)
15. Buffer E (see Recipes)
16. Buffer R (see Recipes)
Recipes
1. Adenine stock (5 mg/mL)
Autoclave at 120 °C for 20 min.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Adenine sulfate | 27.1 mM (in adenine base) | 0.5 g |
| H2O (deionized) | n/a | 99.75 mL |
2. YPD agar plate
Autoclave at 120 °C for 20 min. Pour the medium into a cell culture dish (approximately 20 mL/dish). Solidify the plates for 2–3 days before use, wrap in plastic wrap, and store at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Yeast extract | 1% (w/v) | 10 g |
| Bacto-peptone | 2% (w/v) | 20 g |
| D(+)-glucose | 2% (w/v) | 20 g |
| Agar | 2% (w/v) | 20 g |
| H2O (deionized) | n/a | 1 L |
3. YPDA
Add the adenine after autoclaving at 120 °C for 20 min.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Yeast extract | 1% (w/v) | 10 g |
| Bacto-peptone | 2% (w/v) | 20 g |
| D(+)-glucose | 2% (w/v) | 20 g |
| H2O (deionized) | n/a | 1 L |
| 5 mg/mL adenine stock | 0.02 mg/mL | 4.02 mL |
4. 10× EB (20 mL)
Store at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 1.5 M | 1.75 g |
| 0.5 M EDTA | 10 mM | 0.4 mL |
| 1 M Tris-HCl pH 7.5 | 400 mM | 8 mL |
| H2O (deionized) | n/a | Up to 20 mL |
5. 0.5 M DTT (20 mL)
Aliquot and store at -20 °C. Avoid repeated freeze-thaw cycles.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| (+/-)-Dithiothreitol | 1 M | 1.54 g |
| H2O (deionized) | n/a | 20 mL |
6. EBD (50 mL)
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10×EB | 1× | 5 mL |
| 1 M DTT | 1 mM | 50 μL |
| H2O (deionized) | n/a | 44.95 mL |
7. 0.1 M PMSF (10 mL)
Caution: Be careful not to inhale the powder or allow it to come into contact with skin or eyes. Store at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PMSF | 0.1 M | 0.174 g |
| Ethanol | n/a | 10 mL |
8. EBDPI (10 mL)
Dissolve the tablet in a 15 mL tube by gently stirring. Store at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EBD | n/a | 10 mL |
| cOmplete protease inhibitor cocktail mini (EDTA-free) | n/a | 1 tablet |
9. EBDPIP (5 mL)
Use within 30 min after preparation.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EBDPIP | n/a | 5 mL |
| 0.1 M PMSF | 1 mM | 50 μL |
10. 1 M MgCl2 (10 mL)
Store at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| MgCl2·6H2O | 1 M | 2.03 g |
| H2O (deionized) | n/a | Up to 10 mL |
11. EBMD (10 mL)
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10×EB | 1× | 1 mL |
| 1 M MgCl2 | 5 mM | 50 μL |
| 0.5 M DTT | 10 mM | 20 μL |
| H2O (deionized) | n/a | Up to 10 mL |
12. EBM10D (2 mL)
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10×EB | 1× | 200 μL |
| 1 M MgCl2 | 5 mM | 10 μL |
| 0.5 M DTT | 10 mM | 40 μL |
| H2O (deionized) | n/a | 1.75 mL |
13. 50% PEG8000 (10 mL)
Mix all components except DTT in a 50 mL tube and incubate in a water bath for 20 min at 45 °C. Invert the tube repeatedly to dissolve the PEG. Allow to cool to room temperature and add DTT.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| PEG8000 | 50% (w/v) | 5 g |
| 10× EB | 1× | 1 mL |
| 1 M MgCl2 | 5 mM | 50 μL |
| 0.5 M DTT | 1 mM | 20 μL |
| H2O (deionized) | n/a | Up to 10 mL |
14. 20% PEG8000 (1 mL)
Pipette the viscous 50% PEG8000 using a 1,000 μL tip with the end cut off and mix with EBMD by repeated pipetting.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 50% PEG8000 | 20% (w/v) | 0.4 mL |
| EBMD | n/a | 0.6 mL |
15. Buffer E (10 mL)
Store at 4 °C. Final composition: 2.5 mM D-desthiobiotin, 0.15 M NaCl, 1 mM EDTA, 100 mM Tris-HCl, 1 mM DTT, pH 8.0.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Buffer E (10×) | 1× | 1 mL |
| 0.5 M DTT | 1 mM | 20 μL |
| H2O (deionized) | n/a | Up to 10 mL |
16. Buffer R (10 mL)
Store at 4 °C. Final composition: 1 mM hydroxy-azophenyl-benzoic acid (HABA), 0.15 M NaCl, 1 mM EDTA, 100 mM Tris-HCl, pH 8.0.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Buffer R (10×) | 1× | 1 mL |
| H2O (deionized) | n/a | Up to 10 mL |
Laboratory supplies
1. Cell culture dish, sterilized (SANSEI MEDICAL CO., LTD., catalog number: 01-013)
2. Pipette tips [Watson® Bio Lab, catalog numbers: 123R-755YS (200 μL), 123R-757CS (1000 μL)]
3. Sterile culture tube (16 mL, polypropylene) (EVERGREEN, catalog number: 222-2393-080)
4. Erlenmeyer flask with baffle, 500 mL (SHIBATA, catalog number: 016310-500A)
5. Centrifuge tube, 50 mL, flat top, polypropylene (TRUELINE, catalog number: TR2005)
6. 2 mL tube with screw cap and O-ring, polypropylene (YASUI KIKAI, catalog number: STC-0250)
7. Glass beads (0.5 mm diameter) (YASUI KIKAI, catalog number: YGBLA05)
8. Cell counting slides for TC10™/TC20™ cell counter, dual chamber (Bio-Rad, catalog number: 1450015)
9. Slide glass (MATSUNAMI, catalog number: SFF-001)
10. Coverslip (No. 1 thickness) (MATSUNAMI, catalog number: C018181)
11. Double-sided tape, 5 mm wide (NICHIBAN, catalog number: NW-5)
12. Pasteur pipette, 150 mm (AS ONE, catalog number: 2-2045-01)
13. Silicon dropper, 1 mL (AS ONE, catalog number: 1-6227-02)
14. Empty gravity flow column (1.1 cm diameter and 4.5 cm height) (Amersham Pharmacia, catalog number: 27-4570-01 or equivalent)
15. Low protein binding tube, 1.5 mL (BMS, catalog number: Dot-LP150)
16. Low protein binding tube, 0.5 mL (BMS, catalog number: Dot-LP050)
17. Low protein binding pipette tip [BMS, catalog number: BC-UP-0110S (10 μL), BC-UP-2010S (200 μL), BC-UP-1010S (1000 μL)]
18. Toothpicks (6.5–18 cm) (any brand)
Note: Sterilize before use.
Equipment
1. Automated cell counter (Bio-Rad, model: TC-20)
2. Hybrid high-speed refrigerated centrifuge (KUBOTA, model: 6200)
3. Tabletop micro refrigerated centrifuge (KUBOTA, model: 3520)
4. Bench-top shaking water bath (TAITEC, model: Personal-11 SDN set)
5. VORTEX-GENIE 2 Mixer (M&S Instruments Inc., model: SI-0286)
6. Multi-beads shocker (YASUI KIKAI, model: MB901)
7. Inverted fluorescent microscope (Nikon, model: Eclipse Ti2-E) equipped with:
a. 100× objective lens (Nikon, model: CFI Plan Apoλ 100× Oil Ph3 DM/NA1.45)
b. High-pressure mercury lamp (Nikon, model: Intensilight C-HGFIE 130 W)
c. CMOS image sensor (Nikon, model: DS-Qi2)
d. FITC filter set (Nikon, model: Ex465-495/DM505/BA515-555)
8. Bench-top aspirator (INTEGRA, model: VACUSIP)
9. Inverted routine microscope (Nikon, Eclipse Ts2) equipped with a 40× objective lens (Nikon, CFI Achromat LWD ADL 40XF/NA 0.55, catalog number: MRP46402)
10. Autoclave (TOMY SEIKO CO., LTD, model: LBS-325)
11. Incubator (TAITEC, model: INCUBATE BOX M-200F)
12. Culture rotator (TAITEC, model: RT-50)
13. Rotary shaker (NISSIN, model: NX-20)
14. Blue LED illuminator (NIHON EIDO, model: NF-1050-B)
15. Dark box for LED illumination (NIHON EIDO, model: NF-1060)
16. Pipettes [BM EQUIPMENT Co., LTD, PipetPAL®, PAL-200 (20–200 μL), PAL-1000 (100–1,000 μL)]
17. Stand with flat table and steel support (AS ONE, catalog number: 6-425-03 or any brand)
18. One-sided opening clamp (AS ONE, catalog number: 6-776-05 or any brand)
19. Silicone tubing (any brand)
20. Paper clips (any brand)
Software and datasets
1. NIS-Element AR (Nikon, v. 4.30.01)
2. Fiji (Fiji contributors [11], v. 2.16.0/1.54g)
3. R programs (The R Foundation for Statistical Computing Platform, v. 4.3.3)
4. RStudio Desktop (Posit Software, v. 2023.09.0+463)
Procedure
文章信息
稿件历史记录
提交日期: Mar 4, 2025
接收日期: May 9, 2025
在线发布日期: May 22, 2025
出版日期: Jun 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Takaine, M. (2025). In vitro Condensation Assay of Fluorescent Protein-Fused PRPP Amidotransferase Purified from Budding Yeast Cells. Bio-protocol 15(11): e5335. DOI: 10.21769/BioProtoc.5335.
分类
生物化学 > 蛋白质 > 自组装
生物化学 > 蛋白质 > 分离和纯化
生物化学 > 蛋白质 > 标记
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link