- EN - English
- CN - 中文
TurboID Labeling and Analysis of Proteins in the Primary Cilium
利用TurboID标记与分析初级纤毛中的蛋白质
发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5303 浏览次数: 3027
评审: Jibin SadasivanAlexandre SmirnovAnonymous reviewer(s)
Abstract
Known as the cell’s antenna and signaling hub, the primary cilium is a hair-like organelle with a few micrometers in length and 200–300 nm in diameter. Due to the small size of the primary cilium, it is technically challenging to profile ciliary proteins from mammalian cells. Traditional methods, such as physical isolation of cilia, are susceptible to contamination from other cellular components. Other proximity-based labeling methods via APEX or BioID have been used to map ciliary proteins. However, these approaches have their inherent limitations, including the use of toxic reagents like H2O2 and prolonged labeling kinetics. Here, we show a new proximity-based labeling technique for primary cilia with TurboID. TurboID presents a distinct advantage over BioID and APEX2 due to its expedited labeling kinetics, taking minutes instead of hours, and its use of a non-toxic biotin substrate, which eliminates the need for H2O2. When targeted to the cilium, TurboID selectively labels ciliary proteins with biotin. The biotinylated proteins are then enriched with streptavidin beads and labeled with tandem mass tags (TMT), followed by mass spectrometry (MS) detection. This protocol eliminates the requirement of toxic labeling reagents and significantly reduces the labeling time, thus providing advantages in mapping signaling proteins with high temporal resolution in live cells.
Key features
• Compared to other proximity labeling enzymes, TurboID offers fast labeling kinetics and uses cell-permeable biotin as the labeling reagent [1].
• This protocol includes a straightforward subcellular fractionation step to remove the nuclei to reduce the non-specific background.
• This protocol has been successfully applied to the NIH 3T3 cell line and could also be applied in other cell lines and animal tissues.
Keywords: Proximity labeling (邻近标记)Graphical overview
Background
The primary cilium is a hair-like organelle that protrudes from the surface of nearly all vertebrate cells. Most cilia are a few micrometers long and 200–300 nm in diameter. The ciliary membrane is approximately 1/1,000 to 1/5,000 of the total cellular surface, and the volume is around 1/30,000 of the total cellular volume [2,3]. Acting as cellular antennae, primary cilia sense a wide variety of extracellular cues and transduce them into intracellular signaling pathways [4,5]. Cilium dysfunction causes a wide range of human diseases, collectively known as ciliopathies. Ciliopathies affect the function of almost all human organs, including the brain, eyes, heart, kidneys, liver, skeleton, and reproductive system [6]. To understand the pathogenesis of ciliopathies, it is crucial to identify and characterize the signaling molecules within cilia.
However, progress along this line has been hindered by technical challenges associated with studying such a minuscule compartment of the cell. Conventional approaches of physically isolating cilia produce inconsistent results due to the significant amounts of contaminations from non-ciliary structures, such as microvilli [7–10]. In addition, some ciliary proteins are present in the cilium at very low abundance [11,12], and most signaling transducers transit through the cilia in a very dynamic manner during signal transduction. Physical isolation is not able to capture this dynamic protein transport into and out of the cilium. Proximity labeling–based approaches, circumventing these challenges, have been used to profile ciliary proteins in several studies [11,13–16]. Two categories of proximity labeling enzymes have been developed and used in proximity labeling. One category is peroxidase, such as APEX/APEX2, which adds biotin-phenol to its substrates in the presence of hydrogen peroxide (H2O2). The other category is biotin ligase, which catalyzes the covalent transfer of biotin to its neighboring proteins within 10–20 nm. Each proximity labeling enzyme has its advantages and inherent limitations. APEX/APEX2 has rapid labeling kinetics within a couple of minutes; however, its substrate biotin-phenol shows poor membrane permeability [17,18]. This limits its application to certain cell types and tissues. Using hydrogen peroxide in the labeling process is also a concern because it may affect the cellular oxidative status and induce stress responses [19]. The first generation of promiscuous biotin ligase, BioID, originates from bacteria. BioID requires a long labeling time (24 h) with biotin [15,16], which restricts its application in mapping the dynamic translocation of ciliary proteins during signal transduction.
The new generation of biotin ligase, TurboID, is an engineered biotin ligase derived from BioID. However, the labeling kinetics of TurboID are significantly improved in comparison with its predecessor. TurboID labels proteins within 5 min. Therefore, TurboID offers a fast labeling speed comparable to APEX/APEX2, while eliminating the need for the toxic labeling reagent hydrogen peroxide. Thus, TurboID is ideal for mapping dynamic protein composition with high temporal resolution in live cells. However, TurboID may exhibit elevated baseline biotinylation due to endogenous biotin, which may lead to increased background labeling concerns when mapping dynamic proteins.
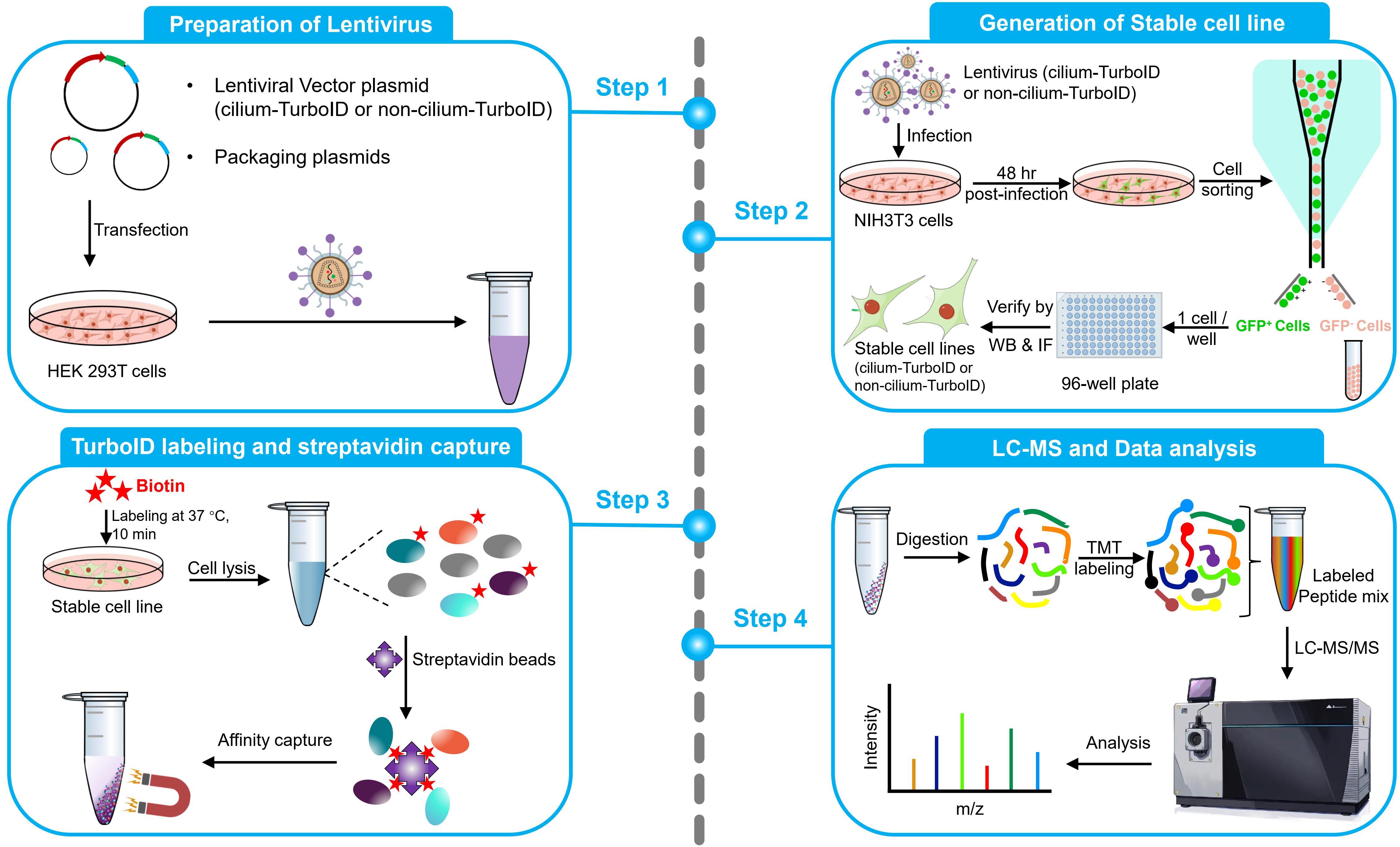
This protocol aims to obtain quantitative cilium proteome with TurboID. It covers the full process of quantitative proteomics, including preparation of lentivirus, generation of a stable cell line, TurboID labeling and streptavidin capture, liquid chromatography–mass spectrometry (LC–MS), and quantitative analysis of mass spectrometry results. To probe the ciliary protein, we fused the TurboID to the ciliary-targeting domain (N+RVEP+PR) and non-ciliary-targeting domain (PR) of the small GTPase Arl13b (cilium-TurboID and non-cilium-TurboID). Our results confirmed the presence of over 100 proteins that were previously reported to localize to the cilium, underscoring its validation in mapping ciliary proteome. Meanwhile, we identified hundreds of new ciliary candidates that can be harvested for further studies [20]. Given that TurboID features a labeling radius of 10–20 nm, which enables precise labeling and identification of proteins close to the bait, leading to the effective identification of bait interactors, this protocol can also be adapted to map protein–protein interactions.
Materials and reagents
Biological materials
1. Bacterial strains: TOP10 (Thermo Fisher Scientific, catalog number: C404010), Stbl3 (Thermo Fisher Scientific, catalog number: C737303)
2. Cell lines: 293T cells (ATCC, catalog number: CRL-3216), NIH 3T3 cells (ATCC, catalog number: CRL-1658)
Reagents
1. psPAX2 plasmid (ampicillin resistance) (Addgene, catalog number: 12260); pMD2.G plasmid (ampicillin resistance) (Addgene, catalog number: 12259); Cilium-TurboID (pFUGW-NRVEPPR-GFP-TurboID, ampicillin resistance) and non-cilium-TurboID (pFUGW-PR-GFP-TurboID, ampicillin resistance) plasmids as described in [20]
2. Ampicillin, sodium salt (Research Products International, catalog number: A40040-5.0)
3. LB broth (Fisher Scientific, catalog number: BP1426-500)
4. Maxi Fast-Ion Plasmid kit (IBI Scientific, catalog number: Ib47122)
5. DMEM (Corning, catalog number: 10-013-CM)
6. Fetal bovine serum (FBS) (Corning, catalog number: 35-010-CV)
7. Penicillin/streptomycin (10,000 U/mL) (Thermo Fisher Scientific, catalog number: 15-140-122)
8. Calcium Phosphate Transfection kit (Takara Bio, catalog number: 631312); includes 2 M calcium solution and 2× HBS
9. Calf serum (CS) (Fisher Scientific, catalog number: 16-010-159)
10. 0.05% trypsin-EDTA (Fisher Scientific, catalog number: 25-300-120)
11. DPBS (Fisher Scientific, catalog number: 14-190-136)
12. Poly-D-lysine (Sigma, catalog number: A003E) (store at -20 °C)
13. Biotin (Sigma, catalog number: B4501-1G)
14. DMSO (Sigma, catalog number: D8418-50ML)
15. 20% PFA (Fisher Scientific, catalog number: 50-980-493)
16. Donkey serum (Sigma, catalog number: S30-100ML)
17. Triton X-100 (Sigma, catalog number: T8787-50ML)
18. HEPES (Fisher Scientific, catalog number: BP310-500)
19. KCI (Fisher Scientific, catalog number: P217-500)
20. MgCl2 (Fisher Scientific, catalog number: AC223211000)
21. 1 M sodium hydroxide (NaOH) (Sigma, catalog number: 1091371003)
22. Hydrochloric acid (HCl) 36.5% to 38.0% (Fisher Scientific, catalog number: A144-500)
23. 0.5 M EDTA (Fisher Scientific, catalog number: AAJ15694AE)
24. 0.5 M EGTA (Fisher Scientific, catalog number: 50-255-956)
25. Tris base (Fisher Scientific, catalog number: BP152-500)
26. NaCl (Fisher Scientific, catalog number: S271-500)
27. SDS (Fisher Scientific, catalog number: BP166-100)
28. Na2CO3 (Fisher Scientific, catalog number: S263-500)
29. Hoechst (Sigma, catalog number: 94403-1ML)
30. Chicken anti-GFP (Aves labs, catalog number: GFP-1020)
31. Mouse anti-Ac-tubulin (Sigma, catalog number: T6793)
32. Donkey anti-chicken AlexaFluor 488 (Jackson ImmunoResearch Labs, catalog number: 703-545-155)
33. Donkey anti-mouse rhodamine (Jackson ImmunoResearch Labs, catalog number: 715-025-151)
34. Alexa Fluor 647 Streptavidin (Jackson ImmunoResearch Labs, catalog number: 016-600-084)
35. Urea (Sigma, catalog number: U5128-500G)
36. Fluoromount-G mounting medium (SouthernBiotech, catalog number: 0100-01)
37. Protease inhibitor cocktail (Roche, catalog number: 11836170001)
38. 10% NP-40 (Thermo Fisher Scientific, catalog number: 28324)
39. BCA Protein Assay kit (Thermo Fisher Scientific, catalog number: 23225)
40. Streptavidin magnetic beads (Thermo Fisher Scientific, catalog number: 88817)
41. 6× Laemmli sample buffer (MP Biomedicals, catalog number: 04822261)
42. Rabbit anti-GFP (Thermo Fisher Scientific, catalog number: A-11122)
43. Mouse anti-GAPDH (Santa Cruz Biotechnology, catalog number: sc-32233)
44. HRP-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Labs, catalog number: 711-035-152)
45. HRP-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Labs, catalog number: 715-035-151)
46. HRP-conjugated streptavidin (Thermo Fisher Scientific, catalog number: N100)
47. 1 M Triethylammonium bicarbonate (TEAB) (Sigma, catalog number: 18597-100ML)
48. 16plex TMT label reagent set (Thermo Fisher Scientific, catalog number: A44521)
49. 0.4% trypan blue solution (Thermo Fisher Scientific, catalog number: 15250061)
Solutions
1. 50 mM biotin stock solution (see Recipes)
2. Biotin labeling medium (see Recipes)
3. Blocking buffer (see Recipes)
4. Subcellular fractionation buffer (see Recipes)
5. 10× TBS buffer (see Recipes)
6. 25× protease inhibitor cocktail buffer (see Recipes)
7. TEAB buffer (see Recipes)
8. 1 M KCl (see Recipes)
9. 0.1 M Na2CO3 (see Recipes)
10. 10 mM Tris-HCl buffer (see Recipes)
11. 2 M urea buffer (see Recipes)
Recipes
1. 50 mM biotin stock solution
Store at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Biotin | 50 mM | 0.5 g |
| DMSO | n/a | 41 mL |
| Total | n/a | 41 mL |
2. Biotin labeling medium
Store at 4 °C.
| Reagent | Final concentration | Volume |
|---|---|---|
| Calf serum (CS) | 0.5% | 250 µL |
| 50 mM biotin stock (Recipe 1) | 50 µM | 50 µL |
| DMEM | n/a | 49.7 mL |
| Total | n/a | 50 mL |
3. Blocking buffer
Store at 4 °C.
| Reagent | Final concentration | Volume |
|---|---|---|
| Donkey serum | 2% | 2 mL |
| Triton X-100 | 0.2% | 200 µL |
| DPBS | n/a | 97.8 mL |
| Total | n/a | 100 mL |
4. Subcellular fractionation buffer
Store at 4°C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 20 mM | 4.77 g |
| KCI | 10 mM | 0.75 g |
| MgCl2 | 2 mM | 0.19 g |
| 0.5 M EDTA | 1 mM | 2 mL |
| 0.5 M EGTA | 1 mM | 2 mL |
| H2O (MilliQ water) | n/a | See note |
| Total | n/a | 1 L |
Note: Allow the reagents to mix completely and adjust the pH of the solution to 7.4 by adding 1 M NaOH while measuring with a pH meter. Then, bring the volume up to 1 L with MilliQ water. Just before use, add and dilute the 25× protease inhibitor cocktail buffer to 1×.
5. 10× TBS buffer
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris | 200 mM | 24.23 g |
| NaCl | 1.5 M | 87.66 g |
| 0.5 M EDTA | 10 mM | 20 mL |
| H2O (MilliQ water) | n/a | See note |
| Total | n/a | 1 L |
Note: Allow the reagents to mix completely and adjust the pH of the solution to 7.4 by adding HCl (36.5% to 38.0%) while measuring with a pH meter. Then, bring the volume up to 1 L with MilliQ water.
6. 25 × protease inhibitor cocktail buffer
Store at -20 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Protease inhibitor cocktail | 25× | 1 tablet |
| H2O (MilliQ water) | n/a | 2 mL |
| Total | n/a | 2 mL |
7. TEAB buffer
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 1 M TEAB | 50 mM | 50 mL |
| NaCl | 150 mM | 8.77 g |
| 0.5 M EDTA | 1 mM | 2 mL |
| H2O (MilliQ water) | n/a | See note |
| Total | n/a | 1 L |
Note: Allow the reagents to mix completely and adjust the pH of the solution to 7.4 by adding HCl (36.5% to 38.0%) while measuring with a pH meter. Then, bring the volume up to 1 L with MilliQ water.
8. 1 M KCl
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| KCl | 1 M | 7.46 g |
| H2O (MilliQ water) | n/a | 100 mL |
| Total | n/a | 100 mL |
9. 0.1 M Na2CO3
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2CO3 | 0.1 M | 1.06 g |
| H2O (MilliQ water) | n/a | 100 mL |
| Total | n/a | 100 mL |
10. 10 mM Tris-HCl buffer
Store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris | 10 mM | 1.21 g |
| H2O (MilliQ water) | n/a | 1 L |
| Total | n/a | 1 L |
Note: Allow the Tris to mix completely and adjust the pH of the solution to 8.0 by adding HCl (36.5% to 38.0%) while measuring with a pH meter. Then, bring the volume up to 1 L with MilliQ water.
11. 2 M urea buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Urea | 2 M | 1.2 g |
| 10 mM Tris-HCl buffer (Recipe 10) | n/a | 10 mL |
| Total | n/a | 10 mL |
Note: Make this solution fresh on the day of use. No need to adjust the pH of the solution.
Laboratory supplies
1. 10 cm dish (Fisher Scientific, catalog number: FB012924)
2. 15 cm dish (Thermo Fisher Scientific, catalog number: 150468)
3. 0.45 µm syringe filter (Fisher Scientific, catalog number: 726-2545)
4. 15 mL conical tube (Corning, catalog number: 430791)
5. 70 µm cell strainer (Falcon, catalog number: 352350)
6. Polystyrene round-bottom tube (Falcon, catalog: 352058)
7. 96-well plate (Thermo Fisher Scientific, catalog number: 12-556-008)
8. 12 mm glass coverslips (Bellco Glass, catalog number: 194310012A)
9. 24-well plate (Fisher Scientific, catalog number: FB012929)
10. Parafilm (Fisher Scientific, catalog number: PM999)
11. Cell scraper (Falcon, catalog number: 353085)
12. 1.5 mL protein LoBind tube (Eppendorf, catalog number: 22431081)
13. 5 mL protein LoBind tube (Eppendorf, catalog number: 30108302)
14. 1 mL syringe (BD Biosciences, catalog number: 309628)
15. 27-gauge needle (BD Biosciences, catalog number: 305136)
16. Tweezers (Fine Science Tools, catalog number: 11252-00)
Equipment
1. Purifier Class II biological safety cabinet (Labconoco, model: 36212/36213 TYPE A2)
2. CO2 incubator (Eppendorf, model: CellXpert C170)
3. Vortex mixer (Fisher Scientific, catalog number: 02-215-414)
4. Centrifuge (Eppendorf, model: 5702R)
5. Cell sorter (BD Biosciences, model: FACSAria III)
6. Fluorescence microscope (Leica, model: DMi8 system)
7. Confocal microscope (Zeiss, model: LSM 880)
8. Sonicator equipped with a microtip, measuring 3/32 inches in diameter (Branson, model: SLPt)
9. Magnetic stand (Thermo Fisher Scientific, catalog number: CS15000)
10. Dry block heater (VWR, catalog number: 75838-282)
11. Savant SpeedVac vacuum concentrator (Thermo Fisher Scientific, model: SPD210-115)
12. UPLC system (Waters, model: ACQUITY UPLC M-Class)
13. Mass spectrometer (Thermo Fisher Scientific, model: Orbitrap Fusion Tribrid MS)
Software and datasets
1. FACSDiva (BD Biosciences, v7.0.x)
2. LAS X (Leica, v3.7.4.23463)
3. ImageJ (NIH, v1.53k)
4. ZEN (Zeiss, ZEN v2.3)
5. Preview (Protein Metrics, v3.5.0.0)
6. Proteome Discoverer (Thermo Fisher Scientific, v1.5)
7. Byonic (Protein Metrics, v3.5.0.0)
Procedure
文章信息
稿件历史记录
提交日期: Jan 29, 2025
接收日期: Apr 11, 2025
在线发布日期: Apr 23, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Liu, X. and Ge, X. (2025). TurboID Labeling and Analysis of Proteins in the Primary Cilium. Bio-protocol 15(9): e5303. DOI: 10.21769/BioProtoc.5303.
分类
细胞生物学 > 细胞结构 > 纤毛
生物化学 > 蛋白质 > 标记
系统生物学 > 蛋白质组学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link