- EN - English
- CN - 中文
Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
利用PISA分析评估代谢物与叶绿体蛋白的相互作用
(§Technical contact: anna.karlsson@scilifelab.se) 发布: 2025年05月05日第15卷第9期 DOI: 10.21769/BioProtoc.5298 浏览次数: 1991
评审: Ritu GuptaVenkatasalam ShanmugabalajiMichael Enos
Abstract
Plants rely on metabolite regulation of proteins to control their metabolism and adapt to environmental changes, but studying these complex interaction networks remains challenging. The proteome integral solubility alteration (PISA) assay, a high-throughput chemoproteomic technique, was originally developed for mammalian systems to investigate drug targets. PISA detects changes in protein stability upon interaction with small molecules, quantified through LC–MS. Here, we present an adapted PISA protocol for Arabidopsis thaliana chloroplasts to identify potential protein interactions with ascorbate. Chloroplasts are extracted using a linear Percoll gradient, treated with multiple ascorbate concentrations, and subjected to heat-induced protein denaturation. Soluble proteins are extracted via ultracentrifugation, and proteome-wide stability changes are quantified using multiplexed LC–MS. We provide instructions for deconvolution of LC–MS spectra and statistical analysis using freely available software. This protocol enables unbiased screening of protein regulation by small molecules in plants without requiring prior knowledge of interaction partners, chemical probe design, or genetic modifications.
Key features
• Optimization of the PISA assay to study protein–ligand interactions in plant chloroplasts, including isolation of chloroplasts.
• Study of regulation on a proteome level, without genetic manipulation or prior knowledge of interaction partners.
• High proteome coverage, low sample requirement, 5-fold reduction of TMT-labeling cost, and short LC–MS analysis time.
• Adaptable to other organisms, such as bacteria, with minor modifications.
Keywords: Chemoproteomics (化学蛋白质组学)Graphical overview
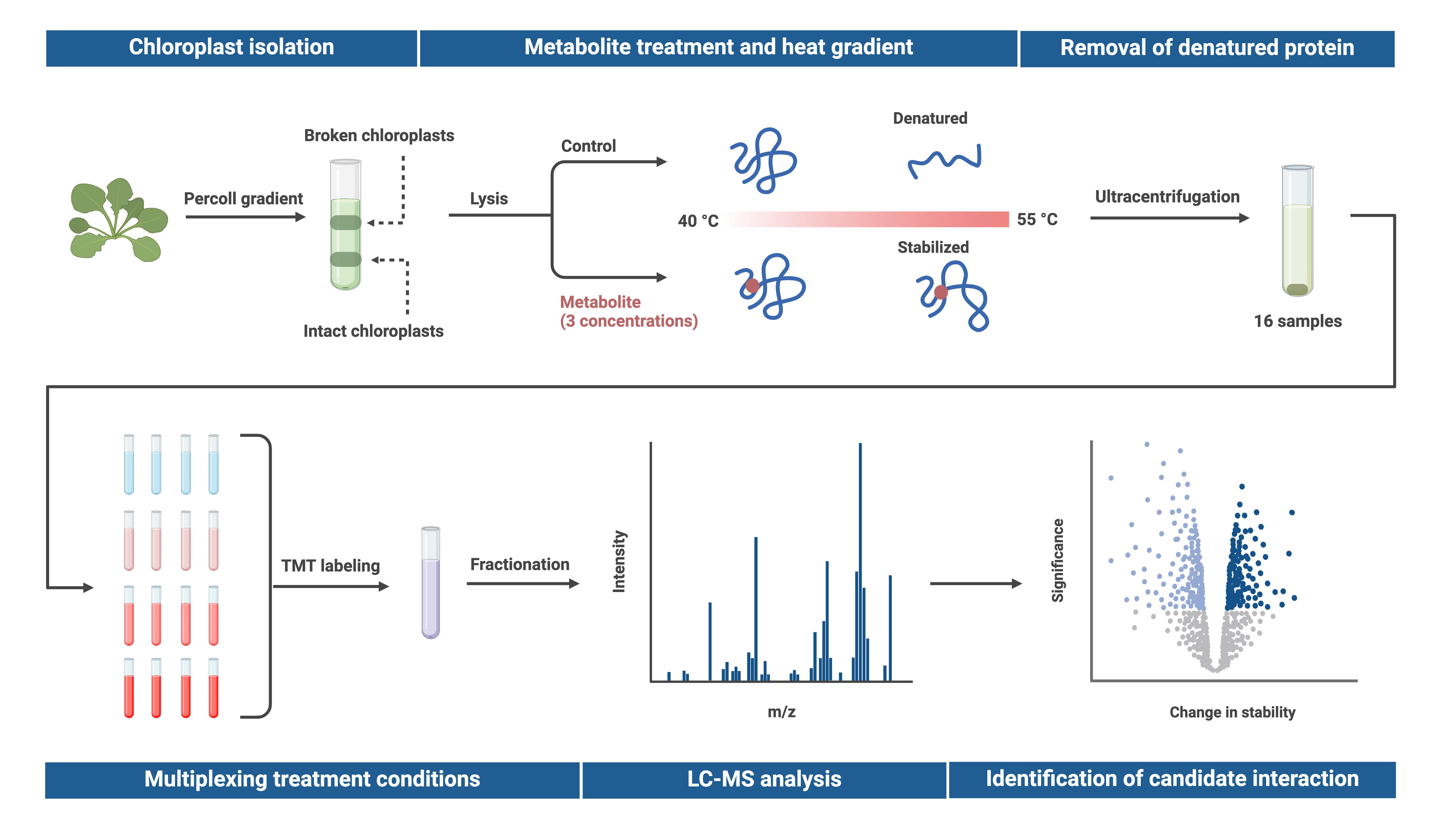
Proteome integral solubility alteration (PISA) assay in plant chloroplasts to screen for protein–metabolite interactions. Intact chloroplasts are isolated from leaves, lysed, and spiked with a metabolite of interest before undergoing temperature treatment. Soluble proteins are extracted via ultracentrifugation, multiplexed using tandem mass tags (TMT), and fractionated. Relative protein abundance is quantified through untargeted proteomics, and changes in protein abundance are compared to a heat-treated control sample without the metabolite. Proteins exhibiting altered temperature stability are considered potential candidates for interaction with the metabolite. To assess dose-response effects, the metabolite is tested at multiple concentrations in technical quadruplicates, with up to three concentrations analyzed using TMTpro 16plex in a single experiment. Created in BioRender. Karlsson, A. (2025) https://BioRender.com/j12o044.
Background
Protein–metabolite interactions create dynamic regulatory networks in plants, regulating basal processes and providing robustness to changes in the environment. Understanding these regulations provides insight into agricultural and biotechnological applications but has historically been understudied due to technological limitations [1–2]. A high-throughput approach for studying protein–metabolite interactions is the proteome integral solubility alteration (PISA) assay, which detects changes in protein solubility upon metabolite binding at a proteome level using LC–MS [3]. The method is based on the principle that metabolite binding can alter protein structure and thereby temperature stability. To assess changes in protein stability, proteome extracts are subjected to a temperature gradient in the presence or absence of a metabolite. By isolating and quantifying the soluble protein fraction, the PISA assay identifies relative changes in protein solubility, indicating potential interactions with the metabolite. The protocol presented here extends the application of the PISA assay from mammalian cells to plant systems and enables the identification of metabolite-interacting proteins in the chloroplast.
The field of proteomics is rapidly advancing, with different approaches to assess protein–metabolite interactions [1–2]. Affinity purification methods use chemically modified or crosslinked metabolites as bait to capture interacting proteins. While effective for identifying high-affinity interactions, the purification process typically disrupts weaker interactions and is prone to higher false-positive rates due to the difficulty of establishing proper controls. Moreover, metabolite modifications can disrupt native interactions and require tailored probe design, limiting the number of candidate metabolites [1]. Alternatively, chemoproteomic methods, such as PISA, detect ligand-induced changes in protein properties without metabolite modification.
Thermal proteome profiling, which PISA is based on, measures protein solubility across multiple temperatures separately to fit a melting curve, determining ligand-induced thermal stability shifts [4]. However, the number of temperature points required for accurate melting curve fitting increases sample consumption and experimental complexity. In contrast, the PISA assay pools individual temperature points after heat treatment and instead quantifies the total solubility change across the temperature gradient. While it does not determine exact melting temperatures, PISA offers higher statistical power, eliminates curve-fitting bias, and significantly reduces sample requirements and mass spectrometry time [3].
In this protocol, we apply the PISA assay to assess potential interactions between chloroplastic proteins and ascorbate, a highly abundant metabolite with diverse functions in plants [5–6]. However, this method can be broadly adapted for screening protein interactions with other metabolites or small molecules, providing a versatile tool for studying plant metabolism. Additionally, the protocol can be extended to other plant compartments or bacteria with minor modifications in protein extraction and temperature gradients, further expanding its applicability to diverse biological contexts.
Materials and reagents
Biological materials
1. Arabidopsis thaliana Col-0 (The European Arabidopsis Stock Centre, catalog number: N1093)
Reagents
1. 2-[4-(2-Hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) (VWR, catalog number: 30487.297)
2. Sorbitol (Merck, catalog number: S1876)
3. Magnesium chloride hexahydrate (Merck, catalog number: M9272)
4. Glycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetic acid (EGTA) (Merck, catalog number: E3889)
5. 2,2',2″,2″'-(Ethane-1,2-diyldinitrilo)tetraacetic acid (EDTA) (Merck, catalog number: E9884)
6. Sodium bicarbonate (Merck, catalog number: S6014)
7. Magnesium sulfate heptahydrate (Merck, catalog number: 1.05886)
8. Percoll (Merck, catalog number: P7828)
9. L-Glutathione reduced (Merck, catalog number: G4251)
10. Potassium chloride (Merck, catalog number: P3911)
11. L-Ascorbic acid (Merck, catalog number: A92902)
12. 3-[4-(2-Hydroxyethyl)piperazin-1-yl]propane-1-sulfonic acid (EPPS) (Thermo Fisher Scientific, catalog number: A13714.14)
13. Dithiothreitol (Fisher Scientific, catalog number: 50-247-531)
14. Iodoacetamide (Merck, catalog number: I1149)
15. Trypsin/Lys-C protease mix (Thermo Fisher Scientific, catalog number: A40009)
16. Hydroxylamine (50% v/v) (Merck, catalog number: 438227)
17. Acetic acid (≥99%) (Merck, catalog number: A6283)
18. Formic acid (≥98%) (Merck, catalog number: 1.00264)
19. Acetonitrile (≥99.9%) (Fisher Scientific, catalog number: 15684740)
20. Trifluoroacetic acid (≥99%) (Merck, catalog number: T6399)
21. Bio-Rad Protein Assay kit (Bio-Rad, catalog number: 5000002)
23. EmporeTM SPE Disks for C18 StageTips (Merck, catalog number: 66883-U)
24. TMTproTM 16plex Label Reagent Set (1 × 0.5 mG) (Thermo Fisher Scientific, catalog number: A44521)
25. PierceTM High pH Reversed-Phase Peptide Fractionation kit (Thermo Scientific, catalog number: 84868)
26. NP-40 Surfact-AmpsTM detergent solution (Thermo Scientific, catalog number: 85125)
27. Potassium hydroxide (KOH) (Merck, catalog number: 484016)
Solutions
1. 1 M HEPES-KOH pH 8 buffer (see Recipes)
2. 2× concentrated isolation buffer (see Recipes)
3. HMS buffer (see Recipes)
4. Percoll gradient (see Recipes)
5. 100 mM HEPES pH 8 buffer (see Recipes)
6. 10× concentrated lysis buffer (see Recipes)
7. High 5× concentrated metabolite solution (see Recipes)
8. Medium 5× concentrated metabolite solution (see Recipes)
9. Low 5× metabolite solution (see Recipes)
10. 20 mM EPPS pH 8.5 buffer (see Recipes)
11. 200 mM dithiothreitol (see Recipes)
12. 375 mM iodoacetamide (see Recipes)
13. Trypsin/Lys-C protease mix (see Recipes)
14. 5% hydroxylamine solution (see Recipes)
15. 0.1% formic acid (see Recipes)
16. 20% formic acid (see Recipes)
17. 80% acetonitrile solution including 0.1% formic acid (see Recipes)
18. 0.1% trifluoroacetic acid (see Recipes)
19. LC-MS solvent A (see Recipes)
20. LC-MS solvent B (see Recipes)
Recipes
1. 1 M HEPES pH 8 buffer
Adjust the final solution to pH 8 using concentrated KOH. Stable for at least 6 months at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 1 M | 23.8 g |
| MilliQ water | n/a | Add to a final volume of 100 mL |
| Total | n/a | 100 mL |
2. 2× concentrated isolation buffer
Prepare fresh one day before the experiment and store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sorbitol | 0.6 M | 109.3 g |
| Magnesium chloride hexahydrate | 10 mM | 2.0 g |
| EGTA | 10 mM | 3.8 g |
| EDTA | 10 mM | 2.9 g |
| Sodium bicarbonate | 20 mM | 1.7 g |
| 1 M HEPES pH 8 buffer (Recipe 1) | 40 mM | 40 mL |
| MilliQ water | n/a | Add to a final volume of 1 L |
| Total | n/a | 1 L |
3. HMS buffer
Prepare fresh one day before the experiment and store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Sorbitol | 0.3 M | 5.5 g |
| Magnesium sulfate heptahydrate | 3 mM | 74 mg |
| 1 M HEPES pH 8 buffer (Recipe 1) | 20 mM | 2 mL |
| MilliQ water | n/a | Add to a final volume of 100 mL |
| Total | n/a | 100 mL |
4. Percoll gradient
Prepare fresh one day before the experiment and store at 4 °C.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Percoll | 50% v/v | 13 mL |
| 2× concentrated isolation buffer (Recipe 2) | 50% v/v | 13 mL |
| Glutathione | 625 µM | 5 mg |
| Total | n/a | 26 mL |
5. 100 mM HEPES pH 8 buffer
Stable for at least 6 months at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HEPES | 100 mM | 1 mL |
| MilliQ water | n/a | 9 mL |
| Total | n/a | 10 mL |
6. 10× concentrated lysis buffer
Stable for at least 6 months at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Magnesium chloride hexahydrate | 30 mM | 61 g |
| Potassium chloride | 1.5 M | 1.1 g |
| 1 M HEPES pH 8 buffer (Recipe 1) | 100 mM | 1 mL |
| MilliQ water | Add to a final volume of 10 mL | |
| Total | n/a | 10 mL |
7. High 5× concentrated metabolite solution
Prepare quickly on ice and shielded from light to avoid degradation. Store at -20 °C in 50 μL PCR tube aliquots. Stable for at least 1 week.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| L-Ascorbic acid | 50 mM | 88 mg |
| 1 M HEPES pH 8 buffer (Recipe 1) | 100 mM | 1 mL |
| MilliQ water | 9 mL | |
| Total | n/a | 10 mL |
8. Medium 5× concentrated metabolite solution
Prepare quickly on ice and shielded from light to avoid degradation. Store at -20 °C in 50 μL PCR tube aliquots. Stable for at least 1 week.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| High 5× concentrated metabolite solution (Recipe 7) | 25 mM | 5 mL |
| 1 M HEPES pH 8 buffer (Recipe 1) | 100 mM | 0.5 mL |
| MilliQ water | 4.5 mL | |
| Total | n/a | 10 mL |
9. Low 5× concentrated metabolite solution
Prepare quickly on ice and shielded from light to avoid degradation. Store at -20 °C in 50 μL PCR tube aliquots. Stable for at least 1 week.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Medium 5× concentrated metabolite solution (Recipe 8) | 12.5 mM | 5 mL |
| 1 M HEPES pH 8 buffer (Recipe 1) | 100 mM | 0.5 mL |
| MilliQ water | 4.5 mL | |
| Total | n/a | 10 mL |
10. 20 mM EPPS pH 8.5 buffer
Adjust the final solution to pH 8.5 using concentrated KOH. Stable for at least 6 months at room temperature.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| EPPS | 20 mM | 51 mg |
| MilliQ water | n/a | 10 mL |
| Total | n/a | 10 mL |
11. 200 mM dithiothreitol
Store at -20 °C in 700 μL single-use aliquots. Stable for at least 6 months.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Dithiothreitol | 200 mM | 154 mg |
| MilliQ water | n/a | 5 mL |
| Total | n/a | 5 mL |
12. 375 mM iodoacetamide
Store at -20 °C in 850 μL single-use aliquots protected from light. Stable for at least 6 months.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Iodoacetamide | 375 mM | 328 mg |
| MilliQ water | n/a | 5 mL |
| Total | n/a | 5 mL |
13. Trypsin/Lys-C protease mix
Store at -20 °C in 25 μL single-use aliquots. Stable for at least 6 months.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetic acid (≥99%) | 0.1% v/v | 0.5 μL |
| MilliQ water | 50% v/v | 500 μL |
| Lyophilized trypsin/lysC mix | 0.2 μg/μL | 100 μg |
| Total | n/a | 500 μL |
14. 5% hydroxylamine solution
Store at 4 °C in an air-tight container for up to 1 week.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Hydroxylamine (50% v/v) | 5% v/v | 100 μL |
| MilliQ water | n/a | 900 μL |
| Total | n/a | 500 μL |
15. 0.1% formic acid
Stable at room temperature in an air-tight container for at least 6 months.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Formic acid (≥98%) | 0.1% v/v | 50 μL |
| MilliQ water | n/a | 50 mL |
| Total | n/a | 50 mL |
16. 20% formic acid
Stable at room temperature in an air-tight container for at least 6 months.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Formic acid (≥98%) | 20% v/v | 2 mL |
| MilliQ water | n/a | 8 mL |
| Total | n/a | 10 mL |
17. 80% acetonitrile solution including 0.1% formic acid
Stable at room temperature in an air-tight container for at least 1 month.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetonitrile (≥99.9%) | 80% v/v | 8 mL |
| 0.1% formic acid (Recipe 14) | 20% v/v | 2 mL |
| Total | n/a | 10 mL |
18. 0.1% trifluoroacetic acid
Stable at room temperature in an air-tight container for at least 3 months.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Trifluoroacetic acid (≥99%) | 0.1% v/v | 50 μL |
| MilliQ water | 99.9% v/v | Add to a final volume of 50 mL |
| Total | n/a | 50 mL |
19. LC–MS solvent A
Degas the solvent for 20 min in an open container using an ultrasonic cleaner. Store in an air-tight container at room temperature and prepare a new solvent every month.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetonitrile (≥99.9%) | 3% v/v | 3 mL |
| Formic acid (>98%) | 0.1% v/v | 100 μL |
| MilliQ water | 99.6% v/v | 997.9 mL |
| Total | n/a | 1 L |
20. LC–MS solvent B
Degas the solvent for 20 min in an open container using an ultrasonic cleaner. Store in an air-tight container at room temperature and prepare a new solvent every month.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Acetonitrile (≥99.9%) | 95% v/v | 950 mL |
| Formic acid (>98%) | 0.1% v/v | 100 μL |
| MilliQ water | 4.9% v/v | 4.9 mL |
| Total | n/a | 1 L |
Laboratory supplies
1. Polycarbonate bottle for Percoll gradient 50 mL (Beckman Coulter, catalog number: 357002)
2. Polycarbonate bottle for centrifugation 500 mL (Beckman Coulter, catalog number: 355664)
3. Polycarbonate tube for ultracentrifugation 1 mL (Beckman Coulter, catalog number: 343778)
4. Miracloth (Millipore, catalog number: 475855)
5. Glass beads 425–600 μm (Sigma-Aldrich, catalog number: G8772)
6. Bead beating 2 mL tubes (Sarstedt Inc, catalog number: 72.694.600)
7. Low protein binding tubes 2 mL (Sarstedt Inc, catalog number: 72.695.600)
8. Screw cap tubes 50 mL (Sarstedt, catalog number: 62.547.254)
9. Screw cap tubes 15 mL (Sarstedt, catalog number: 62.554.502)
10. PCR 8-strip tubes (BRAND, catalog number: 781320)
11. Zeba Spin desalting columns 0.5 mL (Thermo Scientific, catalog number: 89883)
12. V-bottom 96-well plate (Greiner, catalog number: 651201)
13. Clear F-bottom 96-well plate (Greiner, catalog number: 655101)
14. MS vial 1 mL (Waters, catalog number: 186000234)
15. pH indicator strips (Supelco, catalog number: 1.09535)
16. Soft paintbrush for chloroplast resuspension (Princeton Artist Brush Co., catalog number: Series 3050 P3050DF025)
17. Funnel (BRAND, catalog number: 148045)
Equipment
1. MilliQ water purification system (Millipore, model: IQ 7000)
2. Homogenizer (IKA, model: T 25 digital ULTRA-TURRAX)
3. Absorbance plate reader (Molecular Devices, model: SpectraMax i3x)
4. Centrifuge for 2 mL tubes (Eppendorf, model: 5430 R with rotor FA-45-30-11)
5. Centrifuge for 96-well plates and 50 mL tubes (Thermo Fisher Scientific, model: Sorvall ST 16R with rotors M-20 and TX-400)
6. High-speed centrifuge for 50 and 500 mL (Beckman Coulter, model: Avanti J-26 XP with rotors JA-25.50 and JLA 8.1)
7. Ultracentrifuge (Beckman Coulter, model: Optima MAX-XP with rotor TLA 120.2)
8. Thermal cycler with gradient function (Bio-Rad, model: C1000 Touch™ Thermal Cycler with Dual 48/48 Fast Reaction Module)
9. SpeedVac (Genevac, model: miVac Quatro)
10. Thermomixer (Eppendorf, model: F2.0)
11. 6500 K light source (Co/Tech, catalog number: 36-4855)
12. Bead beater (MP Biomedicals, model: FastPrep-24 5G)
13. C18 Acclaim PepMap 100 trap column (Thermo Scientific, catalog number: 164946)
14. ES802 EASY-Spray PepMap RSLC C18 column (Thermo Scientific, catalog number: 164941)
15. Nanoflow UHPLC system (Thermo Fisher Scientific, model: UltiMate 3000 RSLCnano)
16. Mass spectrometer equipped with an EASY ElectroSpray source with MS/MS resolution ≥ 45,000 (Thermo Fisher Scientific, model: Q Exactive HF Hybrid Quadrupole-Orbitrap)
17. Ultrasonic cleaner to degas LC–MS solvents (Branson, model: Bransonic CPX Digital Bath 3800)
18. Growth chamber for plants (Binder, model: KBW 400)
19. PAR light intensity meter (Skye, model: SKP 200 with sensor SKP 217)
20. Multipipette 5–50 μL (Thermo Scientific, model: Finnpipette F1 5–50 μL)
Software and datasets
1. Q Exactive HF Tune program (Thermo Fisher Scientific, version 2.4 or later)
2. Thermo Scientific SII for Xcalibur (Thermo Fisher Scientific, version 1.6 or later)
3. MaxQuant software (Max Planck Institute of Biochemistry, version 2.2.0.0 or later)
4. SoftMax Pro plate reader software (Molecular Devices, version 6.5 or later)
5. R (R Foundation for Statistical Computing, version 4.2.2. or later)
6. Code has been deposited to GitHub: https://github.com/emilsporre/pisa (03/25/2025)
7. MS proteomics raw data used to verify the protocol [5] is available at the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) (dataset identifier: PXD052756)
Procedure
文章信息
稿件历史记录
提交日期: Feb 18, 2025
接收日期: Apr 2, 2025
在线发布日期: Apr 17, 2025
出版日期: May 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Karlsson, A., Sporre, E., Strandberg, L., Tóth, S. Z. and Hudson, E. P. (2025). Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay. Bio-protocol 15(9): e5298. DOI: 10.21769/BioProtoc.5298.
- Tóth, D., Tengölics, R., Aarabi, F., Karlsson, A., Vidal-Meireles, A., Kovács, L., Kuntam, S., Körmöczi, T., Fernie, A. R., Hudson, E. P. et al. (2024). Chloroplastic ascorbate modifies plant metabolism and may act as a metabolite signal regardless of oxidative stress. Plant Physiol. 196(2): 1691–1711. https://doi.org/10.1093/plphys/kiae409
分类
植物科学 > 植物生物化学 > 蛋白质 > 相互作用
系统生物学 > 蛋白质组学
生物化学 > 蛋白质 > 相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link