- EN - English
- CN - 中文
Scalable Alkaline Extraction Protocol for Microbial DNA Screening by PCR
适用于PCR微生物DNA筛查的可扩展碱提取方法
(*contributed equally to this work) 发布: 2025年04月20日第15卷第8期 DOI: 10.21769/BioProtoc.5290 浏览次数: 1709
评审: Alba BlesaAnonymous reviewer(s)

相关实验方案
深海无脊椎动物组织保存的比较方案:DNA/RNA Shield 与液氮在高质量核酸双重提取中的应用对比
Ana S. Gomes [...] Olivier Laroche
2025年11月20日 1319 阅读
Abstract
In molecular diagnosis, DNA extraction kits are sample-specific and proprietary, preventing lateral distribution among similar facilities from different sectors to alleviate supply shortages during a crisis. Previous fast extraction protocols such as detergent-based ones allow fast DNA extraction for nucleic acid amplification tests (NAAT), mainly polymerase chain reaction (PCR). The use of NaOH (dense alkali) to rupture cells and nuclei and destabilize the conformation of DNases might alleviate shortages and costs while retaining enough robustness to treat complicated samples with minimal environmental and logistical footprint. Biological samples are hand-crushed using a pestle in 1.5 mL tubes with 360 μL of 0.2 M NaOH for 3–5 min and incubated at 75 °C for 10 min. For immediate use, 115.2 μL of 1 M Tris (pH 8) and 364.8 μL nuclease-free water are added, and the sample is vortexed for 10 s and spun at 10,000× g for 3 min; then, 700 μL is transferred to a clean microtube. Two serial dilutions follow, and all concentrations are used as templates for PCR. A refined, storable extract can be produced by adding 70 μL of HCl 1 M (instead of Tris-HCl) and one volume of cold isopropanol to the extract for standard precipitation. This method can increase throughput in emergencies by field deployment in resource-limited settings (RLS) or allow benchtop backup in cases of acquisition disruption or sample surge in established facilities. The crude extract can be used for immediate PCR in both benchtop and portable thermocyclers, thus allowing NAAT in resource-limited settings with low costs and waste footprint or during prolonged crises, where supply chain failures may occur. The refined version produces alcohol-precipitated nucleic acids, suitable for both immediate use and for storage or dispatch for spatiotemporally separate analysis while offering much better amplification quality with a small increase in time and minimal increase in expendables/chemicals needed.
Key features
• DNA extraction from different sample types using only boiling water and occasional mechanical assistance.
• Crude extract serially diluted to bypass purification and quantification steps.
• Refined extract is partly purified, more enriched, storable, and transportable and contributes to higher sensitivity.
• Both versions decrease costs and the overall footprint of testing to increase sustainability in field operations and in standard lab environments under supply chain derailment.
Keywords: Alkaline extraction (碱提取)Graphical overview
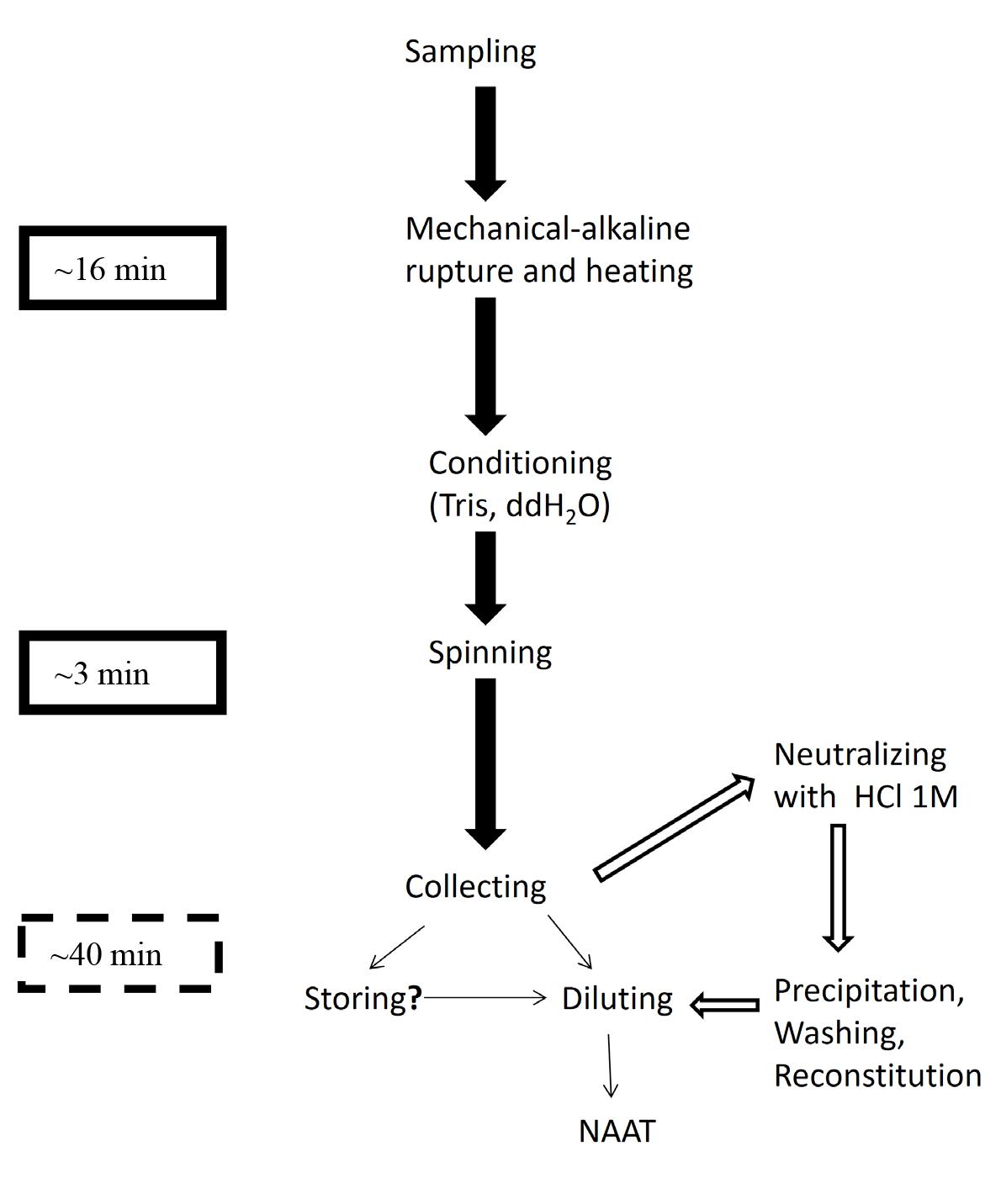
Successive steps of the scalable NaOH-based extraction protocol. Solid arrows and text box: Short protocol, crude extract. Hollow arrows and dashed text box: Refinements. Line arrows: Post-extraction procedures: Procedure not tested.
Background
In routine conditions or in crisis, surveillance and screening require fast and massive DNA extraction pipelines for nucleic acid amplification tests (NAAT), especially PCR, both in field conditions [usually in resource-limited settings (RLS)] and in proper lab setups. Although current technology for DNA extraction achieves significant yields from complex and/or small samples, their use is expensive and subject to market manipulation and practices and supply chain integrity [1,2]. The alkaline approach has been suggested for contingency [3] and field conditions [4]. It is a straightforward and inexpensive method, less susceptible to supply chain and market fluctuations as it uses only widely available, inexpensive chemicals and expendables. In particular, NaOH—the main lysis factor—is much more efficient than ultra-short, detergent-based approaches, either as kits or as household formulations [5] or boiling water [6]. Due to its scalability, which is its novelty, the protocol described herein may result in a template for immediate use in NAAT (mainly PCR) or in an extract that is storable, transportable, much more enriched, and amplification-friendly. In both cases, the serial dilutions just before use overcome the lack of a spectrophotometer in RLS to manage concentration uncertainties [7].
In a health or ecological crisis, massive spatial dispersion and increased processing output of incoming samples may overload and overrun standard public health or agronomic facilities and infrastructures. Thus, ease of procurement and use by personnel moderately trained in molecular diagnosis but experienced in other sectors of biosciences may increase the processing rate in point-of-collection (PoC) settings, thus neutralizing infectious potential and offloading work from dedicated infrastructures; the latter would tackle unresolved or suspect samples and provide confirmatory analyses as needed. The choice for in situ refinement allows safe dispatching of samples to distant facilities [8], which is otherwise expensive, creates biosafety and biosecurity risks, and may degrade the samples, thus devaluing the actual diagnostic procedure.
The present protocol may be used in RLS, ideally combined with portable instrumentation [9] in its crude, fast version for immediate testing in time-sensitive scenarios, and/or severe RLS. In more permissive scenarios and conditions, its refined version may be implemented, producing much better results [3]. This protocol may be used to improve the turnaround time and volume throughput in established, indoor facilities, especially in its refined version, to increase sensitivity. This comes at the expense of other performance metrics, including speed and simplicity. This method may be used for public health and environmental and agronomic emergencies [10], thus qualifying for implementing the One-Health framework. Processing uncomplicated human/animal samples for genotyping genomic biomarkers by basic PCR protocols or other NAAT should be relatively straightforward and requires no modification, as such samples are easier than the ones tested in the validation publication. Blood is always an unknown quantity regarding extraction efficiency and robustness; less demanding samples, such as mouth swabs, are easy to process and sufficient for targeted biomarker analyses since they are within the capability limits of the less robust thermo-osmotic protocol [6]. Additionally, simplex, single-locus PCR results may be further analyzed downstream by restriction digestion [11,12], single-locus sequencing [13], or any other approach.
Prospective operators should be aware of some limitations of this protocol. Namely, the extract is impure, and thus quantification by standard methods is inapplicable. For the same reason, DNA may be degraded due to a number of factors directly affecting its duration (and thus some of them alleviated in the refined version); the same stands for inhibitor molecules that may hamper the NAAT step. The use of different mechanical rupture techniques, such as sonication, is always a possibility but is not included in this protocol. Enzymatic cell lysis may also be tried; however, it is possible that the chemical environment, due to the NaOH, may degrade or nullify the enzymes used, and thus extensive modification may be required. In any case, the use of enzymes clearly undermines the lean, minimal footprint, supply-chain resilient, and affordable nature of this protocol as presented herein and has not been tested.
Materials and reagents
Note: The provisions below do not include expendables and instrumentation necessary for PCR and other NAAT nor for downstream procedures, including but not limited to gel electrophoresis.
Biological materials
1. Klebsiella pneumoniae and Staphylococcus aureus (kindly supplied as anonymous cultures by Assoc Prof. Kolonitsiou Fevronia from the Department of Microbiology, Medical School, University of Patras)
2. Fungal strains of Cryptococcus neoformans, Candida albicans, Aspergillus fumigatus, and Malassezia pachydermatis (all on solid culture on Petri dishes upon identification by NCPF/UoA)
3. Household fruits (tomatoes) showing observable mycelial development, collected by the authors and kept unidentified
4. Petioles excised using a sterile scalpel blade from leaves of olive trees suspect for Xylella spp. (especially X. fastidiosa) infection, collected by the authors (PPDP)
Reagents
1. NaOH, pellets (Riedel-de-Haen, catalog number: 30620)
2. Tris-HCl, powder (Invitrogen, catalog number: 15504-020)
3. Hydrochloric acid (HCl) 37% (PanReac AppliChem, catalog number: 131020.1212)
4. Isopropanol/2-propanol (Sigma-Aldrich, catalog number: 34863)
5. Ethanol (absolute) (Fisher Scientific, catalog number: E/0650DF/17)
6. Water, molecular biology (PanReac AppliChem, catalog number: A7398.100)
Solutions
1. Ethanol 70% v/v (see Recipes)
2. HCl 1 M (see Recipes)
3. Tris-HCl 1 M, pH 8 (see Recipes)
4. NaOH 0.2 M (see Recipes)
Recipes
1. Ethanol 70% v/v
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanol (absolute) | 70% | 700 mL |
| Distilled H2O | 30% | 300 mL |
| Total | n/a | 1,000 mL |
2. HCl 1 M (pH 8)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| HCl 37% | 1 M | 5 mL |
| Distilled H2O | n/a | 45 mL |
| Total | n/a | 50 mL |
Caution: Diluting the commercially available HCl stock solution to working concentrations requires proper precautions due to its vapors and extreme corrosiveness. Use only rated glassware, add the acid slowly in water (not the opposite), and work with gloves (latex is the absolute minimum, but chemical-resistant gloves are the best choice) and other available personal protective equipment (PPE) including but not limited to eye protection and face mask (even standard surgical ones are better than nothing). Also, work only in designated spaces/fume hoods.
3. Tris-HCl 1 M, pH 8
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris-HCl | 1 M | 12.114 g |
| Distilled H2O | n/a | 100 mL |
| Total | n/a | 100 mL |
4. NaOH 0.2 M
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaOH | 0.2 M | 0.8 g |
| Distilled H2O | n/a | 100 mL |
| Total | n/a | 100 mL |
See Troubleshooting 2.
Caution: Solubilizing the commercially available NaOH form (pellets) to working concentrations requires precautions due to the extremely caustic nature of the pellets. Do not make contact with exposed skin and add to the solvent gradually, some pellets at a time. Use only properly rated glassware. Work with gloves (latex is the absolute minimum, but chemical-resistant gloves are preferable) and other available PPE including but not limited to eye protection and face mask (even standard surgical ones are better than nothing). Also, work only in designated spaces/fume hoods.
Laboratory supplies
1. Plastic pellet pestles, blue polypropylene autoclavable (Sigma-Aldrich, catalog number: Z359947-100EA)
2. Scalpel blades (no. 22 stainless disposable; FEATHER, lot 03067430) or box cutter (e.g., Q-Connect, catalog number: 233471)
3. Beaker, 1,000 mL (Hamed, catalog number: 303173)
4. Falcon-shaped plastic tubes, 50 mL (KIMA Vacutest, catalog number: KIMA-17102-50)
5. Plastic microtubes w/cap, 1.5 mL (PierceTM, catalog number: 69715)
6. Pipette tips, 0–200 μL (KIMA Vacutest, catalog number: KIMA-18260)
7. Pipette tips, 1,000 μL (KIMA Vacutest, catalog number: KIMA-18172)
8. Disposable inoculation loops 10 μL, sterile (LABSOLUTE®, catalog number: 7696430)
9. Rack (50-position for 18 mm diameter microtubes) (Fruugo)
Equipment
1. Thermo shaker (Kisker Biotech GmbH & Co, model: TS-100)
2. Pipette P20, 0–20 μL (Gilson PIPETMAN®, catalog number: F123600)
3. Pipette P200, 20–200 μL (Gilson PIPETMAN®, catalog number: F123601)
4. Pipette P1000, 200–1,000 mL (Gilson PIPETMAN®, catalog number: F123602)
Procedure
文章信息
稿件历史记录
提交日期: Feb 5, 2025
接收日期: Mar 30, 2025
在线发布日期: Apr 10, 2025
出版日期: Apr 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Goudoudaki, S., Kambouris, M. E., Kritikou, S., Milioni, A., Velegraki, A., Manoussopoulos, Y. and Patrinos, G. P. (2025). Scalable Alkaline Extraction Protocol for Microbial DNA Screening by PCR. Bio-protocol 15(8): e5290. DOI: 10.21769/BioProtoc.5290.
分类
微生物学 > 病原体检测 > PCR
分子生物学 > DNA > DNA 提取
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link