- EN - English
- CN - 中文
Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
基于Intein的生物传感器用于体内蛋白稳定性监测
(*contributed equally to this work) 发布: 2025年04月20日第15卷第8期 DOI: 10.21769/BioProtoc.5271 浏览次数: 1571
评审: Philipp A.M. SchmidpeterSneha RayAnonymous reviewer(s)
Abstract
Inteins are elements translated within host proteins and removed via a unique protein splicing reaction. In this process, the two peptide bonds flanking the intein are rearranged, releasing the intein and leaving a standard peptide bond in its place. Due to their ability to shuffle peptide bonds in a specific and controlled manner, inteins have proven valuable in protein engineering, leading to the development of numerous impactful technologies. In one application, intein-based biosensors link the activity of a host protein to intein excision. Recently, we developed a biosensor to measure protein stability in vivo, in which the removal of an intein-protein fusion is required for antibiotic resistance. In our protocol, cells expressing our biosensor are logarithmically diluted and spotted on agar plates containing increasing levels of antibiotics. Following incubation, quantitative survival curves can be generated. We also developed a dual protein stability sensor where both antibiotic resistance and fluorescence can be used as readouts and demonstrated that co-expression of the chaperonin GroEL can promote survival and fluorescence. Taken together, our novel intein-based biosensor adds to the available tools to measure protein stability within the cellular environment.
Key features
• Biosensor demonstrates a 100,000-fold difference in survival between stable and unstable test proteins.
• Dual biosensor quantitatively links protein stability to antibiotic resistance and fluorescence simultaneously.
Keywords: Intein (Intein)Background
Inteins (intervening proteins) are polypeptide segments translated within host proteins that are removed through a protein splicing reaction [1,2]. In the intein-mediated splicing reaction, the intein is released by the formation of a peptide bond between the flanking sequences, termed N- and C-exteins, which form the mature host protein. This reaction can proceed without any external factors, though the rate at which splicing occurs is highly dependent on the conditions present. The unique ability of inteins to rearrange peptide bonds has led to the invention of numerous applications in protein engineering [2,3]. Abundant and widespread within the microbial world [4], prior work has demonstrated that some inteins have adapted to serve as environmental sensors, displaying their capacity to regulate protein function in response to conditions such as metals, heat, salt, oxidative stress, and DNA damage [2,5,6].
To monitor intein activity in response to different conditions, protein splicing reporters are often used. These reporters are flexible in design, as their only requirement is the presence of a cysteine, serine, or threonine as the first residue following the intein [1]. Previously, we developed a kanamycin intein splicing reporter (KISR), with it providing an easy method to detect selective small molecule inhibitors of protein splicing within the cellular environment [7–10]. As inteins are absent in humans but present within human pathogens including Mycobacterium tuberculosis and Cryptococcus neoformans, splicing inhibitors may serve as novel antimicrobials, making them increasingly important targets to study [11,12]. Kanamycin A is a commonly used aminoglycoside antibiotic that inhibits translation. Bacteria with a mature aminoglycoside phosphotransferase (KanR), however, can survive in the presence of the antibiotic. The KISR reporter makes use of KanR as the exteins flanking an intein, making it so intein splicing activity confers antibiotic resistance to the experimental cells transformed with the reporter [7–10]. In the presence of small molecule inhibitors, the KISR will be unable to splice, leaving the cells vulnerable to the antibiotic. Therefore, this reporter allows for easy observation of what molecules can act as inhibitors and to what degree they inhibit, as stronger inhibitors will lead to reduced growth.
We recently developed an intein-based biosensor that adapts the KISR framework into a tripartite folding biosensor, where a protein of interest (POI) is inserted within the intein between conserved splicing blocks (Figure 1). Tripartite protein folding biosensors serve to measure the stability of a POI by linking it to reporter activity within the cellular environment [13]. Our intein-based tripartite biosensor design, while serving a similar purpose, distinguishes itself from previous biosensors through the insertion of the POI into an intein, which is then flanked with reporter extein sequences [14]. Through splicing activity, the POI is removed as the extein sequences are joined into a functional reporter molecule. This allows the biosensor to avoid the challenges associated with previous designs that kept a POI fused to the reporter, such as limitations on the size of POI insertion or restrictions on reporter function. Initial work with our biosensor was carried out using the E. coli maltose binding protein (MBP) as a POI, with it being inserted into the homing endonuclease (HEN) loop of the Pyrococcus horikoshii RadA intein (RadA-I) [14]. The HEN loop of RadA-I has been noted to accommodate POI insertion, as previous work has shown that the intein remains splicing active upon insertion of ZsGreen into the loop [15,16]. The RadA-I intein was also selected due to its rapid and accurate splicing across a variety of extein contexts [17–19], which indicated that it could accommodate reporter exteins. The RadA-I-MBP fusion was inserted within KanR (KanR-RadA-I-MBP), with kanamycin resistance serving as the reporter in a manner similar to KISR. This biosensor was then tested in a variety of conditions, where it was observed that a decrease in MBP stability (due to mutations) or inhibition of splicing activity resulted in reduced survival of cells, as they were not provided kanamycin resistance [14]. This supported the viability of our intein-based biosensor approach, as the stability of the POI was directly linked to reporter activity. Further work, which made use of the fluorescent protein TagRFP675 as the POI, showed the potential of our biosensor design as a dual-folding sensor, with both antibiotic resistance and fluorescence linked to POI stability [14]. Potential applications of our intein-based biosensor are broad due to the relaxed requirements of intein splicing, making this approach viable for linking protein stability to any desired in vivo readout [14].
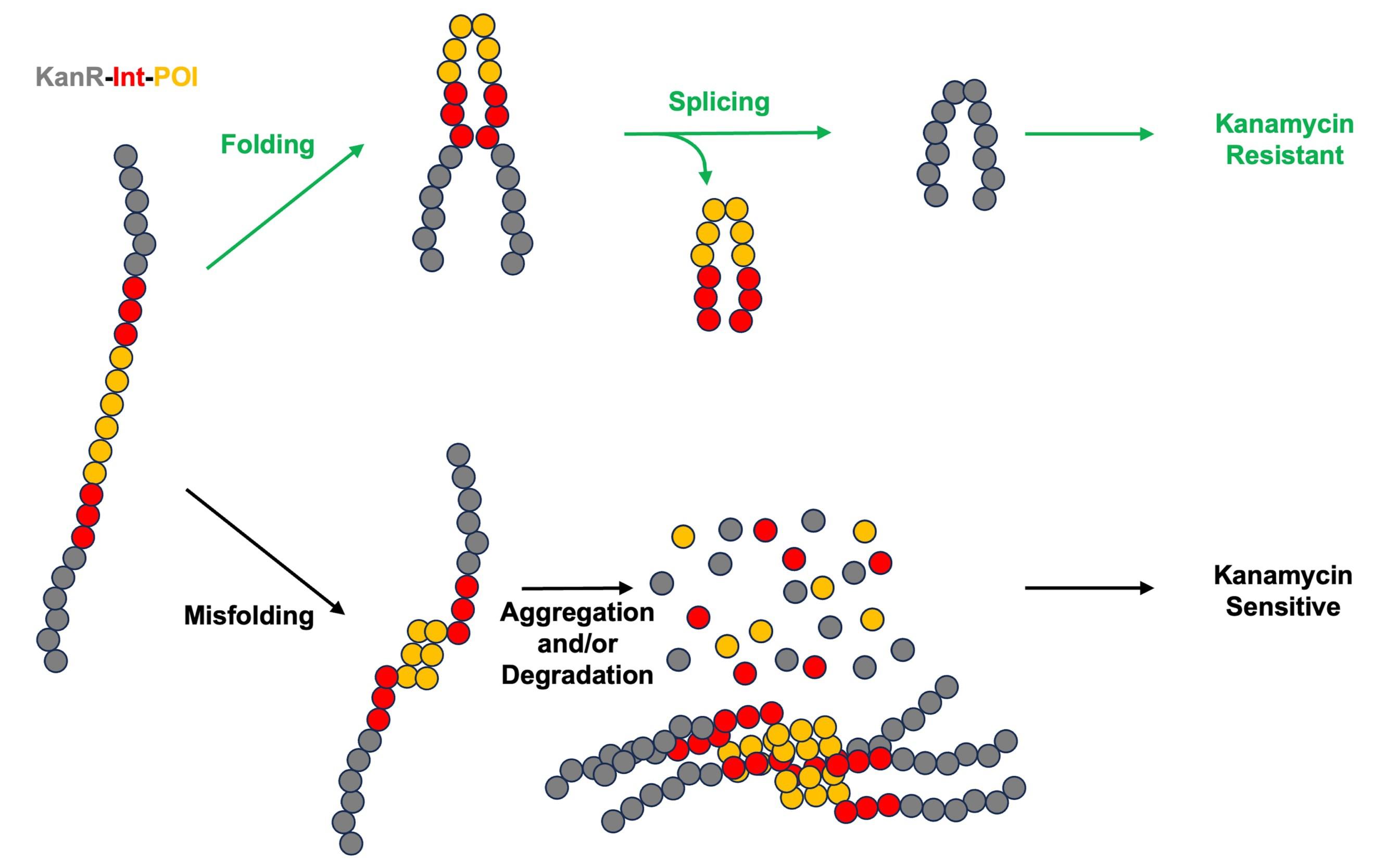
Figure 1. Intein-based protein stability sensor. KanR is colored in gray, intein is colored in red, and the protein of interest (POI) is colored in orange.
Materials and reagents
Biological materials
1. NEB5α E. coli (New England Biolabs, C2987H) (Table 1)
2. MC4100(DE3) [20]
Table 1. E. coli strains used in this study.
| Strain number | Plasmid 1 | Plasmid 2 | Strain | ||
|---|---|---|---|---|---|
| CWL599 | KanR-IntWT-MBPMUT | ApR | NEB5a | ||
| CWL600 | KanR-IntWT-MBPWT | ApR | NEB5a | ||
| CWL640 | KanR-IntMUT-MBPWT | ApR | NEB5a | ||
| AS917 | KanR-IntWT-MBPMUT | ApR | pBAD33mut-Empty | CmR | MC4100(DE3) |
| AS923 | KanR-IntWT-MBPMUT | ApR | pBAD33-GroEL | CmR | MC4100(DE3) |
| AS925 | KanR-IntWT-MBPWT | ApR | pBAD33mut-Empty | CmR | MC4100(DE3) |
| AS931 | KanR-IntWT-MBPWT | ApR | pBAD33-GroEL | CmR | MC4100(DE3) |
| AS933 | KanR-IntWT-TagRFP675 | ApR | pBAD33mut-Empty | CmR | MC4100(DE3) |
| AS939 | KanR-IntWT-TagRFP675 | ApR | pBAD33-GroEL | CmR | MC4100(DE3) |
Reagents
1. Difco LB broth, Miller (BD, catalog number: 244610)
2. Difco LB agar, Miller (BD, catalog number: 244510)
3. Ampicillin sodium salt (Fisher, catalog number: BP906)
4. Kanamycin sulfate (Fisher, catalog number: BP1760)
5. Chloramphenicol (Fisher, catalog number: BP904100)
6. Ethanol (Sigma-Aldrich, catalog number: E7023)
7. L-Arabinose (Chem-IMPEX, catalog number: 29750)
8. Phosphate-buffered saline (PBS) (10×) pH 7.4, RNase-free (Fisher, catalog number: AM9625)
Solutions
1. LB medium (see Recipes)
2. LB agar (see Recipes)
3. 100 mg/mL ampicillin stock solution (1,000×) (see Recipes)
4. 50 mg/mL kanamycin stock solution (1,000×) (see Recipes)
5. 35 mg/mL chloramphenicol stock solution (1,000×) (see Recipes)
6. 20% L-arabinose stock solution (100×) (see Recipes)
Recipes
1. LB medium
25 g of LB broth
Bring to 1 L with ddH2O
Autoclave to sterilize
2. LB agar
40 g of LB agar
Bring to 1 L with ddH2O
Autoclave to sterilize
3. 100 mg/mL ampicillin stock solution (1,000×)
1 g of ampicillin sodium salt
Bring to 10 mL with ultrapure H2O
Place in 10 mL syringe and pass through 0.22 mm filter to sterilize
Aliquot in 1.5 mL tubes and store at -20 °C
4. 50 mg/mL kanamycin stock solution (1,000×)
500 mg of kanamycin sulfate
Bring to 10 mL with ultrapure H2O
Place in 10 mL syringe and pass through 0.22 mm filter to sterilize
Aliquot in 1.5 mL tubes and store at -20 °C
5. 35 mg/mL chloramphenicol stock solution (1,000×)
350 mg of chloramphenicol
Bring to 10 mL with ethanol
Place in 10 mL syringe and pass through 0.22 mm filter to sterilize
Aliquot in 1.5 mL tubes and store at -20 °C
6. 20% L-arabinose stock solution (100×)
2 g of L-arabinose
Bring to 10 mL with ultrapure H2O
Place in 10 mL syringe and pass through 0.22 mm filter to sterilize
Aliquot in 1.5 mL tubes and store at -20 °C
Laboratory supplies
1. 1, 10, 20, 200, and 1,000 μL pipette tips (USA Scientific, Inc. TipOne, catalog numbers: 1111-3800, 1120-1810, 1111-0706, and 1111-2820)
2. 15 mL conical tubes (Greiner Bio-One, catalog number: 188 271)
3. Tissue culture plate 96 wells (Fisher Scientific, catalog number: FB012931)
4. Nonbinding 96-well plates (Greiner Bio-One, catalog number: 655904)
5. Black 96-well plates, black (Corning, catalog number: NBS3991)
6. Petri dishes (Fisher Scientific, catalog number: FB0875712)
7. 1.5 mL cuvette (Fisher Scientific, catalog number: 14955127)
8. 1.5 mL microcentrifuge tubes (USA Scientific, catalog number: 1615-5500)
9. 10 mL syringe (BD, catalog number: 302995)
10. 0.22 mm syringe filters (Fisher Scientific, catalog number: 09-719A)
11. Roll & GrowTM spherical glass plating beads (MP Biomedicals, catalog number: 115000550)
12. Cell scraper (CELLTREAT, catalog number: 229310)
Equipment
1. Benchtop refrigerated shaking incubator (Thermo Scientific, catalog number: SHKE4000)
2. Spectrophotometer (Eppendorf, catalog number: 6131 02779)
3. Amersham Imager 600 (GE, catalog number: 29-0834-61)
4. Microplate reader (Tecan, model: Infinite M200 Pro)
5. P20 Pipetman (Gilson, catalog number: FA10003M)
6. P200 Pipetman (Gilson, catalog number: F144058M)
7. P1000 Pipetman (Gilson, catalog number: FA10006M)
8. Multichannel pipette (Thermo Scientific, catalog number: 46300100)
9. Incubator (Fisher Scientific, model: 11-690-637D)
10. Infrared thermometer (Etekcity, model: 817915020807)
11. Bunsen burner
12. 1 cm grid paper
Procedure
文章信息
稿件历史记录
提交日期: Feb 5, 2025
接收日期: Mar 11, 2025
在线发布日期: Mar 25, 2025
出版日期: Apr 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Smetana, J. S., Ariagno, T. M., Son, A., Horowitz, S. and Lennon, C. W. (2025). Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor. Bio-protocol 15(8): e5271. DOI: 10.21769/BioProtoc.5271.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 活性
分子生物学 > 蛋白质 > 抗微生物分析
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link