- EN - English
- CN - 中文
An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
基于活性蛋白质组学和二维聚丙烯酰胺凝胶电泳(2D-PAGE)鉴定拟南芥细胞间隙液中的靶蛋白酶
发布: 2025年03月05日第15卷第5期 DOI: 10.21769/BioProtoc.5226 浏览次数: 1966
评审: Min CaoAnonymous reviewer(s)

相关实验方案
利用SP3珠和稳定同位素质谱技术优化蛋白质合成速率:植物核糖体的案例研究
Dione Gentry-Torfer [...] Federico Martinez-Seidel
2024年05月05日 2852 阅读
Abstract
Plant proteases participate in a wide variety of biological processes, including development, growth, and defense. To date, numerous proteases have been functionally identified through genetic studies. However, redundancy among certain proteases can obscure their roles, as single-gene loss-of-function mutants often exhibit no discernible phenotype, limiting identification through genetic approaches. Here, we describe an efficient system for the identification of target proteases that cleave specific substrates in the Arabidopsis apoplastic fluid. The method involves using Arabidopsis-submerged culture medium, which contains apoplastic proteases, followed by native two-dimensional electrophoresis. Gel fractionation and an in-gel peptide cleavage assay with a fluorescence-quenching peptide substrate are then used to detect specific proteolytic activity. The active fraction is then subjected to mass spectrometry–based proteomics to identify the protease of interest. This method allows for the efficient and comprehensive identification of proteases with specific substrate cleavage activities in the apoplast.
Key features
• Targets Arabidopsis thaliana secreted protease but may be applicable to other plant species and intracellular proteases if protease-enriched samples are available.
• The protocol involves an in-gel peptide cleavage assay of native two-dimensional gels diced with SAINOME plates, using a fluorescence-quenching substrate.
• Facilitates the efficient identification of proteases with the desired activity from the entire sample, without restricting the analysis to a specific class of proteases.
Keywords: Extracellular protease (细胞外蛋白酶)Graphical overview
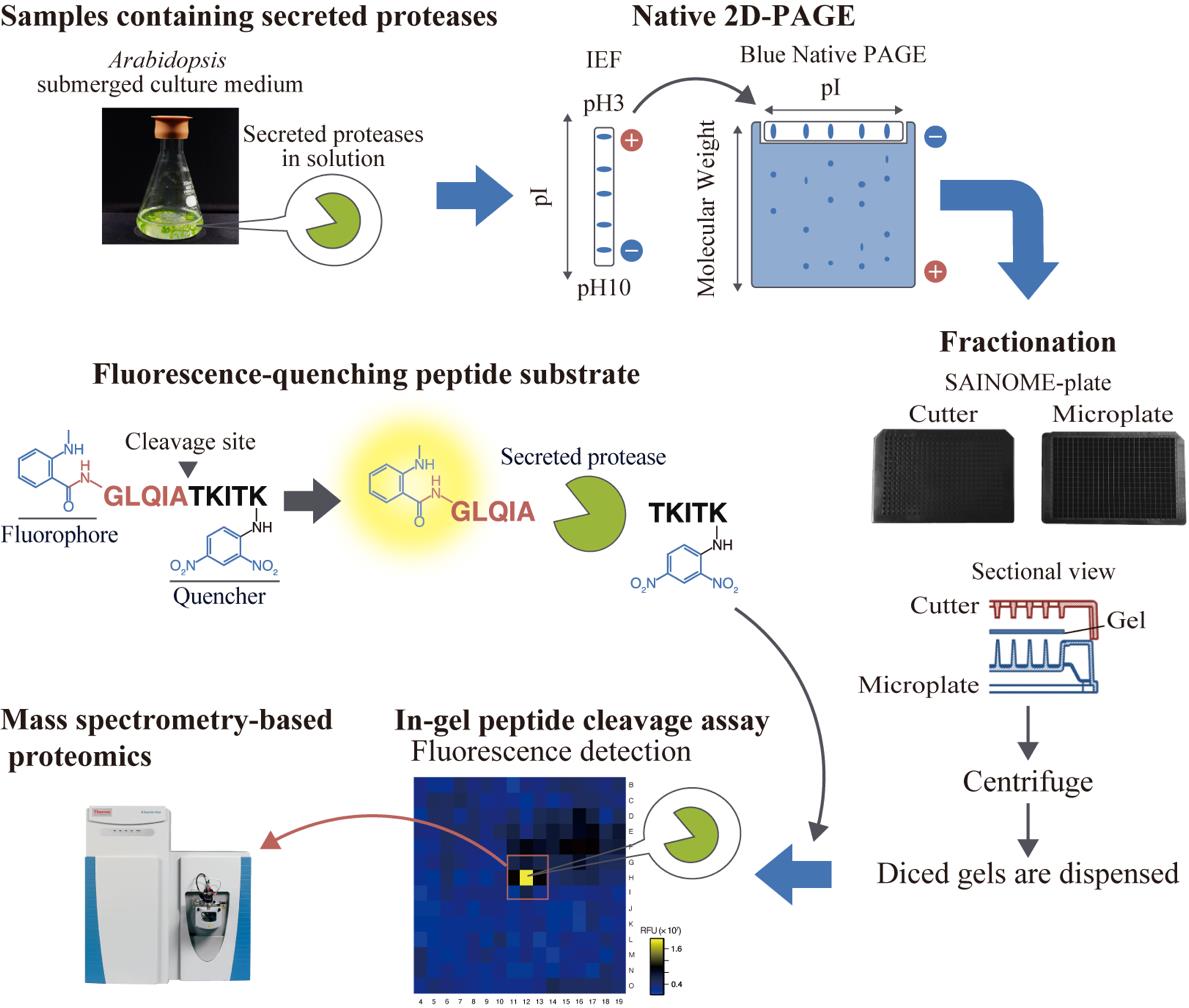
High-throughput system for identifying proteases that cleave specific substrates in Arabidopsis apoplastic fluid. Arabidopsis-submerged culture medium containing secreted proteases is subjected to native two-dimensional electrophoresis. After electrophoresis, the gel is fractionated using a SAINOME plate, and an in-gel peptide cleavage assay is performed using a fluorescence-quenching peptide substrate that fluorescently detects proteolytic activity at the specific cleavage site. Mass spectrometry–based proteomics is performed to identify proteases in the active fraction.
Background
Plant proteases participate in a variety of physiological processes, including development, growth, and defense [1]. Their roles extend beyond protein degradation to include protein activation and regulation of protein localization through limited and site-specific proteolysis [2]. Among the numerous proteases, secreted proteases in the apoplast have received increasing attention in recent years, mainly because of their roles in immunity and peptide hormone processing [3–5]. In the model plant Arabidopsis thaliana, more than 800 putative protease genes have been identified, nearly 300 of which encode secreted proteases with signal peptides [1,6,7].
Proteases often exhibit functional redundancy, which complicates their study through genetic approaches. To address this issue, some studies have employed inhibitors that specifically target particular protease families and suppress their function [8–11]. Alternatively, strategies focusing on changes in transcript levels or enzyme activity under specific conditions have been used to identify proteases of interest [12,13]. Protease identification has also been attempted by narrowing down candidate proteases based on the similarity between the cleavage site of a target substrate and the cleavage sequence of a known protease [14,15]. However, these approaches are all indirect and do not directly assess the specificity of the recognition sequence of the enzyme. Consequently, these methods are not always effective for identifying the protease responsible for cleaving a specific substrate protein.
We have developed a high-throughput system for identifying proteases involved in the cleavage of specific substrates at defined cleavage sites from samples containing apoplastic proteases. This approach employs native two-dimensional (2D) electrophoresis, followed by gel dicing using a SAINOME plate and an in-gel peptide cleavage assay with a fluorescence-quenching peptide substrate [16–19]. Using this system, we recently identified secreted proteases involved in the C-terminal cleavage of the immunogenic peptide flg22, derived from the bacterial flagellar protein flagellin [20]. Here, we provide a step-by-step method for identifying secreted proteases that cleave specific substrates at defined sites, using the identification of flagellin-cleaving proteases as an example. This method is broadly applicable to any proteases that retain enzymatic activity following native 2D electrophoresis.
Materials and reagents
Biological materials
1. Arabidopsis thaliana accession Col-0 submerged culture medium [20] (see General note 1)
Reagents
1. Gamborg's B5 medium salt mixture (Shiotani M.S., catalog number: 399-00621)
2. Sucrose (Wako, catalog number: 199-00027)
3. Nicotinic acid (Nacalai Tesque, catalog number: 24326-52)
4. Pyridoxine hydrochloride (Wako, catalog number: 165-05401)
5. Thiamin hydrochloride (Wako, catalog number: 201-00852)
6. Myo-inositol (Sigma-Aldrich, catalog number: I5125)
7. Glycine (Wako, catalog number: 077-00735)
8. Agar (Wako, catalog number: 016-11875)
9. Spectra/Por 7 membrane (molecular weight cutoff 10 kDa) (Repligen, catalog number: 132117)
10. 3-[(3-Cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) (Dojindo, catalog number: C008)
11. Immobilized pH gradient (IPG) buffer, pH 3–10 NL (GE Healthcare, catalog number: 17-6000-88)
12. (+/-)-Dithiothreitol (DTT) (Wako, catalog number: 049-08972)
13. Bromophenol blue (BPB) (Wako, catalog number: 021-02911)
14. Immobiline DryStrip pH 3–10 NL, 7 cm (IPG strip) (Cytiva, catalog number: 17-6001-12)
15. Immobiline DryStrip cover fluid (Cytiva, catalog number: 17-1335-01)
16. 6-Aminocaproic acid (LKT Laboratories, catalog number: A4935)
17. Glycerol (Nacalai Tesque, catalog number: 17018-25)
18. 2-Amino-2-hydroxymethyl-1,3-propanediol (Tris) (Wako, catalog number: 204-07885)
19. Coomassie brilliant blue G 250 (CBB G-250) (Fluka, catalog number: 27815)
20. NativePAGE running buffer (20×) (Invitrogen, catalog number: BN2001)
21. NativePAGE 4%–16%, Bis-Tris, 1.0 mm, mini protein gels, 15-well (Invitrogen, catalog number: BN1004BOX)
22. NativeMarkTM unstained protein standard (Invitrogen, catalog number: LC0725)
23. Nma-flg[47–56]-Dnp [fluorescence-quenching substrate, Nma-GLQIATKITK(Dnp)-NH2] (Peptide Institute, custom-made) (see General Note 2)
24. 2-Morpholinoethanesulfonic acid, monohydrate (MES) (Dojindo, catalog number: GB12)
25. Methanol (Wako, catalog number: 131-01826)
26. Acetic acid (Wako, catalog number: 017-00256)
27. Lysyl endopeptidase (Lys-C) (Wako, catalog number: 125-05061)
28. Trypsin (Promega, catalog number: V5111)
Solutions
1. B5 vitamin solution (100×) (see Recipes)
2. B5 medium (see Recipes)
3. Rehydration solution A (see Recipes)
4. Rehydration solution B (see Recipes)
5. Rehydration solution (see Recipes)
6. Equilibration solution (see Recipes)
7. Native PAGE anode buffer (see Recipes)
8. Native PAGE cathode buffer (see Recipes)
9. In-gel peptide cleavage assay solution (see Recipes)
10. Fixing solution (see Recipes)
Recipes
1. B5 vitamin solution (100×)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Nicotinic acid | 50 mg/L | 25 mg |
| Pyridoxine hydrochloride | 50 mg/L | 25 mg |
| Thiamin hydrochloride | 10 mg/L | 5 mg |
| Myo-inositol | 10 g/L | 5 g |
| Glycine | 200 mg/L | 100 mg |
| Milli-Q water | n/a | Up to 500 mL |
| Total | n/a | 500 mL |
2. B5 medium
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Gamborg's B5 medium salt mixture | n/a | One bag (3.3 g) |
| B5 vitamin solution (100×) | 1× | 10 mL |
| Sucrose | 1% (w/v) | 10 g |
| Milli-Q water | n/a | Up to 1 L |
| Total | n/a | 1 L |
Adjust pH to 5.7 with 1 N KOH. Autoclave at 121 °C for 20 min. For solid medium, add 7 g of agar before autoclaving.
3. Rehydration solution A
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CHAPS | 4% (w/v) | 40 mg |
| IPG buffer | 0.5% (v/v) | 5 μL |
| DTT (15.9 mg/mL) | 0.62 mg/mL | 39 μL |
| Milli-Q water | n/a | Up to 1 mL |
| Total | n/a | 1 mL |
4. Rehydration solution B
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| CHAPS | 2% (w/v) | 20 mg |
| IPG buffer | 0.5% (v/v) | 5 μL |
| BPB [0.05% (w/v)] | 0.002% (w/v) | 40 μL |
| Milli-Q water | n/a | Up to 1 mL |
| Total | n/a | 1 mL |
5. Rehydration solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Rehydration solution A | 12.5% (v/v) | 20 μL |
| Rehydration solution B | 87.5% (v/v) | 140 μL |
| Total | n/a | 160 μL |
6. Equilibration solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 6-Aminocaproic acid | 450 mM | 0.59 g |
| Glycerol | 25.5% (v/v) | 2.55 mL |
| Tris-HCl (0.5 M, pH 6.8) | 50 mM | 1 mL |
| CBB G-250 | 0.02% (w/v) | 2 mg |
| Milli-Q water | n/a | Up to 10 mL |
| Total | n/a | 10 mL |
7. Native PAGE anode buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 20× Native PAGE running buffer | 1× | 10 mL |
| Milli-Q water | n/a | Up to 200 mL |
| Total | n/a | 200 mL |
8. Native PAGE cathode buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 20× Native PAGE running buffer | 1× | 5 mL |
| CBB G-250 [0.4% (w/v)] | 0.002% (w/v) | 500 μL |
| Milli-Q water | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
9. In-gel peptide cleavage assay solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Nma-flg[47–56]-Dnp (0.4 mM) | 40 μM | 1.7 mL |
| MES-KOH (pH5.7) | 25 mM | 850 μL |
| Milli-Q water | n/a | 14.45 mL |
| Total | n/a | 17 mL |
10. Fixing solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Methanol | 20% (v/v) | 20 mL |
| Acetic acid | 7.5% (v/v) | 7.5 mL |
| Milli-Q water | n/a | 72.5 mL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 300 mL flask (Iwaki, catalog number: 4980FK300)
2. 15 mL centrifuge tubes (Corning, catalog number: 430791)
3. 1.5 mL microtubes (e.g., BMBio, catalog number: BM-15F)
4. Stainless steel surgical blade, No. 14 (FEATHER Safety Razor, model: No. 14)
5. Tweezers (e.g., rubisTech, catalog number: SS-SA)
6. Square Petri dish (Eiken Chemical, catalog number: AW2000)
7. SAINOME, black [SAINOME consists of a microplate (384-well) and a cutter] (Sainome, catalog number: SAI-386-B)
Equipment
1. Rotary vacuum evaporator (EYELA, model: N-N series)
2. High-speed micro centrifuge CF15RN (Hitachi Koki, model: himac CF15RN)
3. Angle rotor (Hitachi Koki, model: T11A31)
4. Angle rotor (Hitachi Koki, model: T15AP31)
5. Strip holder, 7 cm (Cytiva, catalog number: 80-6416-87)
6. Ettan IPGphor II isoelectric focusing unit (Amersham Biosciences, catalog number: 80-6505-03)
7. Mini-slab electrophoretic apparatus (Bio Craft, model: BE-211)
8. High-speed micro centrifuge CF16RN (Hitachi Koki, model: himac CF16RN)
9. Swing rotor (Koki Holdings, model: T5S32)
10. VICTOR Nivo multimode plate reader equipped with a 340/20 nm excitation filter and a 435/20 nm emission filter (PerkinElmer, catalog number: HH35000500)
11. 340/20 nm filter (PerkinElmer, catalog number: HH35000912)
12. 435/20 nm filter (PerkinElmer, catalog number: HH35000918)
13. Extrahera (Biotage, catalog number: 414001)
14. Mass spectrometer (Thermo Fisher Scientific, model: Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer)
15. Gradient pump (Thermo Fisher Scientific, model: Dionex U3000)
Software and datasets
1. VICTOR Nivo software ver. 4.0 (PerkinElmer, 2020 May)
2. Proteome Discoverer ver. 2.3 (Thermo Fisher Scientific)
Procedure
文章信息
稿件历史记录
提交日期: Dec 9, 2024
接收日期: Jan 25, 2025
在线发布日期: Feb 16, 2025
出版日期: Mar 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Matsui, S. and Matsubayashi, Y. (2025). An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid. Bio-protocol 15(5): e5226. DOI: 10.21769/BioProtoc.5226.
分类
植物科学 > 植物生物化学 > 蛋白质 > 分离和纯化
生物化学 > 蛋白质 > 电泳
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link