- EN - English
- CN - 中文
Quantification of Neuromuscular Junctions in Zebrafish Cranial Muscles
斑马鱼头部肌肉神经肌肉接头的定量分析
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5219 浏览次数: 1650
评审: Ivonne SehringAmr Galal Abdelraheem IbrahimSalim Gasmi
Abstract
Communication between motor neurons and muscles is established by specialized synaptic connections known as neuromuscular junctions (NMJs). Altered morphology or numbers of NMJs in the developing muscles can indicate a disease phenotype. The distribution and count of NMJs have been studied in the context of several developmental disorders in different model organisms, including zebrafish. While most of these studies involved manual counting of NMJs, a few of them employed image analysis software for automated quantification. However, these studies were primarily restricted to the trunk musculature of zebrafish. These trunk muscles have a simple and reiterated anatomy, but the cranial musculoskeletal system is much more complex. Here, we describe a stepwise protocol for the visualization and quantification of NMJs in the ventral cranial muscles of zebrafish larvae. We have used a combination of existing ImageJ plugins to develop this methodology, aiming for reproducibility and precision. The protocol allows us to analyze a specific set of cranial muscles by choosing an area of interest. Using background subtraction, pixel intensity thresholding, and watershed algorithm, the images are segmented. The binary images are then used for NMJ quantification using the Analyze Particles tool. This protocol is cost-effective because, unlike other licensed image analyzers, ImageJ is open-source and available free of cost.
Key features
• Immunostaining neuromuscular junctions in alcohol-exposed zebrafish.
• Quantification of presynaptic and postsynaptic terminals in cranial muscles of zebrafish larvae.
• Analysis of size and distribution of NMJs in cranial muscles of zebrafish larvae.
Keywords: Neuromuscular junctions (神经肌肉接头)Graphical overview
Background
In vertebrates, the muscles of the face and neck are innervated by cranial motor neurons [1]. Communication between the brain and muscle is mediated through a specialized synapse known as the neuromuscular junction (NMJ). NMJs typically comprise three components: 1) the axonal end of the motor neuron, forming the presynaptic terminal; 2) the muscle endplate, forming the postsynaptic terminal; and 3) a synaptic cleft that lies between these two terminals. Neuromuscular communication occurs when acetylcholine, released from the presynaptic terminal into the synaptic cleft, binds acetylcholine receptors clustered in the postsynaptic terminal [2]. Alteration in NMJ morphology or distribution can lead to reduced synaptic transmission and result in neuromuscular disease. Zebrafish has emerged as an excellent model to study neuromuscular pathologies [3]. However, characterization and quantification of NMJs in the cranial muscles of zebrafish can be particularly challenging due to the anatomical complexity of the head.
Previous studies involving the quantification of NMJs in the cranial muscles were primarily carried out manually. Manual counting is arduous and can lead to individual bias and subjectivity. Some recent studies have carried out automated quantification using image analysis tools. However, most of these studies were carried out in somites of the zebrafish trunk region [4,5]. Unlike the head, the muscles of the trunk have a simpler, reiterated anatomy. Another study employed Imaris software for surface rendering with automatic thresholding to characterize the NMJs in the cranial musculature [6]. This method is useful for studying the number, shape, and architecture of NMJs. However, this also requires buying licensed software, making it inaccessible to some labs. Also, most of these studies use auto-thresholding, which can change the cutoff value for thresholding in individual images, unintentionally introducing errors [7].
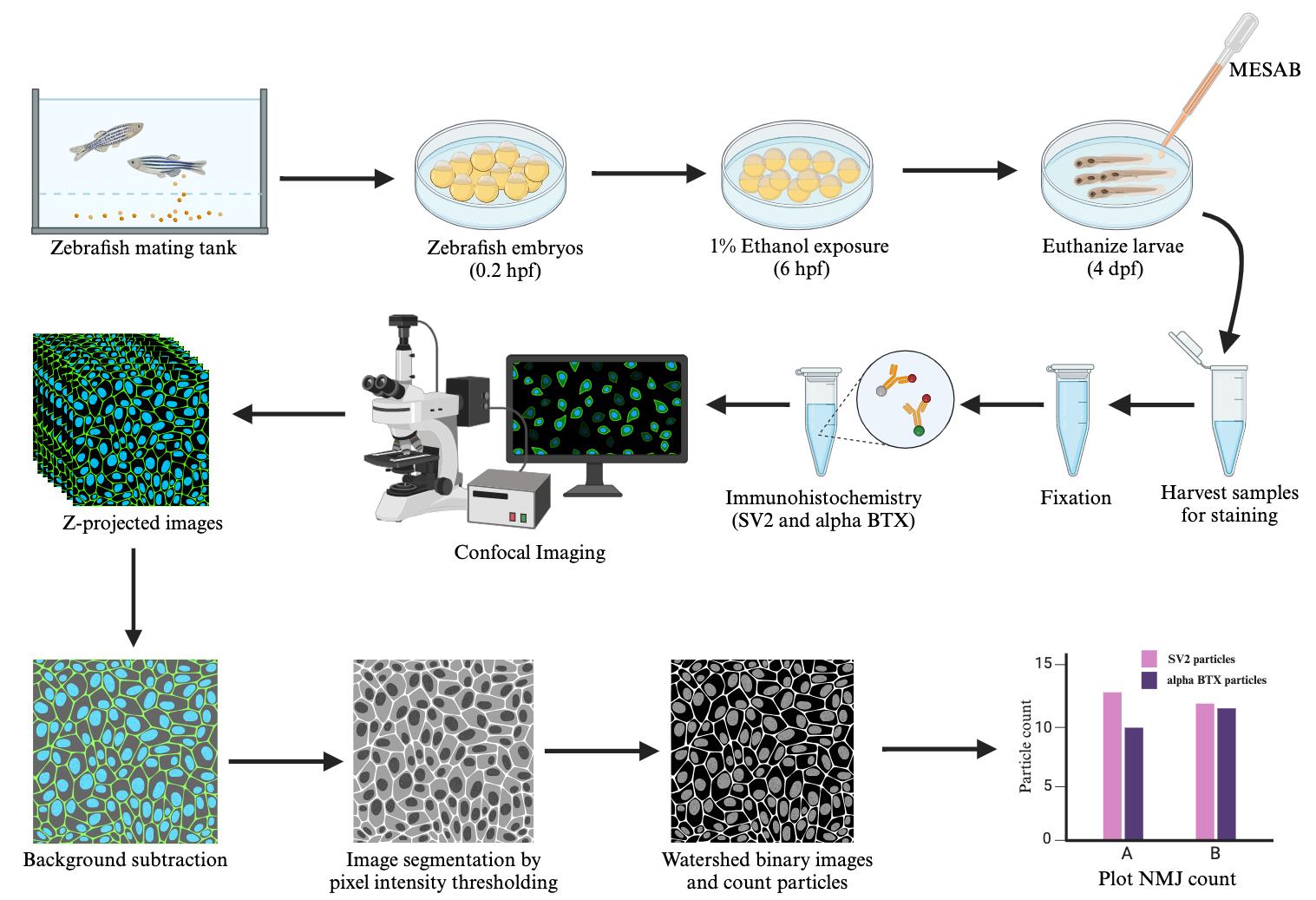
Here, we developed a protocol for NMJ visualization and quantification in the cranial muscles using FIJI/ImageJ software, which is free of cost [8]. We immunolabel the presynaptic and postsynaptic terminals using SV2 antibody and alpha-bungarotoxin, respectively. We then capture high-resolution images of NMJs using confocal microscopy. Using the Area Selection tool, we crop out a region of specific dimensions containing the muscles of interest. We then use the Rolling Ball Background Subtraction plugin, Global Thresholding, and Watershed plugin to segment the images. Instead of auto-thresholding, we set the same cutoff value for thresholding each image, ensuring uniformity across all images. The number of particles in the segmented images is then counted using the Analyze Particles tool. We have used this protocol to investigate the effect of alcohol on NMJ count in a specific set of cranial muscles. However, the method can be applied to other cranial and trunk muscles of zebrafish. The method can also be used to quantify NMJs in other organisms like chickens and mice. Additionally, we used this method with 20× magnification for NMJ counting, but the method could readily be adapted for analyzing images of other magnifications. This method is broadly applicable to quantify other fluorescently labeled particles such as apoptotic cells, proliferating cells, or cells in general by counting DAPI-stained nuclei.
Materials and reagents
Biological materials
1. Zebrafish (Animal husbandry, Eberhart Lab, University of Texas at Austin, USA)
Reagents
1. Alpha-bungarotoxin (α-BTX) Alexa Fluor 647 conjugated (Invitrogen, catalog number: B35450)
2. Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A4503, CAS: 9048-46-8)
3. Calcium chloride dihydrate (CaCl2·2H2O) (Sigma, catalog number: C8106, CAS: 10035-04-8)
4. Dimethyl sulfoxide (DMSO) (EMD Millipore Corp., catalog number: 317275, CAS: 67-68-5)
5. Ethanol (PHARMO, Ethyl Alcohol 100%, catalog number: 111000200, CAS: 64-17-5)
6. Goat anti-mouse IgG, Alexa Fluor 488 (secondary antibody) (Invitrogen, catalog number: A-11029)
7. Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391, CAS: 10034-99-8)
8. MESAB/Tricaine methanesulfonate (Syndel, SYNCAINE/MS 222, FDA approved)
9. Methanol (Fisher Chemical, catalog number: A412-4, CAS: 67-56-1)
10. Methylcellulose (Sigma-Aldrich, catalog number: M0387, CAS: 9004-67-5)
11. Normal goat serum (NGS) (Jackson ImmunoResearch Laboratories, catalog number: 005-000-121)
12. Paraformaldehyde (PFA) (Alfa Aesar, catalog number: A11313, CAS: 30525-89-4)
13. Phosphate-buffered saline (PBS) (Sigma-Aldrich, catalog number: P5368-10PAK)
14. Potassium chloride (KCl) (Fisher chemicals, catalog number: P217, CAS: 7447-40-7)
15. Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P3786, CAS: 7758-11-4)
16. Sodium bicarbonate (NaHCO3) (Sigma-Aldrich, catalog number: S5761, CAS: 144-55-8)
17. Sodium chloride (NaCl) (CHEM-IMPEX INT’L INC., catalog number: 30070, CAS: 7647-14-5)
18. Sodium phosphate dibasic (Na2HPO4) (Acros organics, catalog number: 42437, CAS: 7558-79-4)
19. Synaptic vesicle 2A (SV2) antibody (Developmental Studies Hybridoma Bank, Antibody Registry ID: AB_2315387)
20. Triton X-100 (Triton X) (MP Biomedicals, catalog number: 194854, CAS: 9002-93-1)
Solutions
1. 10× PBS (See Recipes)
2. 1× PBT (See Recipes)
3. 4% PFA (See Recipes)
4. 25% methanol (See Recipes)
5. 50% methanol (See Recipes)
6. 75% methanol (See Recipes)
7. 20× embryo medium stock (See Recipes)
8. 500× sodium bicarbonate (See Recipes)
9. Embryo medium (See Recipes)
10. 1% ethanol (See Recipes)
11. Incubation buffer (See Recipes)
12. Blocking buffer (See Recipes)
13. Ethyl 3 Aminobenzoate methyl sulfonate salt (MESAB)/tricaine solution (See Recipes)
Recipes
1. 10× PBS
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Phosphate-buffered saline | 10× | 1 packet |
| ddH2O | n/a | Up to 100 mL |
| Total | n/a | 100 mL |
2. 1× PBT
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10× PBS | 1× | 5 mL |
| Triton X | 0.5% (v/v) | 250 µL |
| ddH2O | n/a | Up to 50 mL |
| Total | n/a | 50 mL |
3. 4% PFA
*Note: Add PFA to 400 mL of ddH2O and heat and stir in a fume hood until the solution clears (do not heat at more than 60 °C). Add 50 mL of 10× PBS and adjust the pH to 7.4. Add ddH2O up to a total of 500 mL.
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10× PBS | 1× | 50 mL |
| PFA | 4% (w/v) | 20 g |
| ddH2O | n/a | 450 mL* |
| Total | n/a | 500 mL |
Store 4% PFA at -20 °C.
4. 25% methanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Methanol | 25% (v/v) | 1 mL |
| 1× PBT | 75% (v/v) | 3 mL |
| Total | n/a | 4 mL |
5. 50% methanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Methanol | 50% (v/v) | 2 mL |
| 1× PBT | 50% (v/v) | 2 mL |
| Total | n/a | 4 mL |
6. 75% Methanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Methanol | 75% (v/v) | 3 mL |
| 1× PBT | 25% (v/v) | 1 mL |
| Total | n/a | 4 mL |
7. 20× embryo medium stock
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaCl | 0.3 M | 17.5 g |
| KCl | 10.06 mM | 0.75 g |
| CaCl2·2H2O | 19.72 mM | 2.9 g |
| K2HPO4 | 2.35 mM | 0.41 g |
| Na2HPO4 | 1 mM | 0.142 g |
| MgSO4·7H2O | 19.88 mM | 4.9 g |
| ddH2O | n/a | Up to 1 L |
| Total | n/a | 1 L |
Filter-sterilize the solution.
8. 500× sodium bicarbonate
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| NaHCO3 | 0.35 M | 0.3 g |
| ddH2O | n/a | 10 mL |
9. Embryo medium
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 20× embryo medium stock | 1× | 50 mL |
| 500× sodium bicarbonate | 1× | 2 mL |
| ddH2O | n/a | 948 mL |
| Total | n/a | 1 L |
10. 1% Ethanol
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Ethanol | 1% (v/v) | 400 µL |
| Embryo medium | 99% (v/v) | 39.6 mL |
| Total | n/a | 40 mL |
11. Incubation buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 10× PBS | 1× | 5 mL |
| BSA | 1% (w/v) | 500 mg |
| DMSO | 1% (v/v) | 500 µL |
| Triton X-100 | 0.5% (v/v) | 250 µL |
| ddH2O | n/a | Up to 50 mL |
| Total | n/a | 50 mL |
12. Blocking buffer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Incubation buffer | 96% (v/v) | 960 µL |
| Normal goat serum | 4% (v/v) | 40 µL |
| Total | n/a | 1,000 µL |
13. MESAB/tricaine solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Na2HPO4 | 0.8 % (w/v) | 4 g |
| MESAB salt | 0.4 % (w/v) | 2 g |
| ddH2O | n/a | Up to 500 mL |
| Total | n/a | 500 mL |
Adjust pH between 7.0 and 7.2.
Laboratory supplies
1. Petri dishes 100 mm × 20 mm (Falcon, catalog number: 353003)
2. Micropipettes (Sigma-Aldrich, catalog number: FA10006M-1EA)
3. Pipette pumps (Fisher Scientific, catalog number: 13-683C)
4. 1.7 mL microcentrifuge tubes (Fisher Scientific, catalog number: 07-200-535)
Equipment
1. Confocal microscope (Zeiss, model: LSM 980)
2. Stereoscope (Leica, model: KL 300 LED)
Software and datasets
1. ImageJ2 v2.14.0/1.54f (available free of cost from https://fiji.sc/, 07/07/2023)
2. Prism v9.5.0 (GraphPad, 12/06/2022)
Procedure
文章信息
稿件历史记录
提交日期: Oct 15, 2024
接收日期: Jan 2, 2025
在线发布日期: Jan 26, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Ghosal, R. and Eberhart, J. K. (2025). Quantification of Neuromuscular Junctions in Zebrafish Cranial Muscles. Bio-protocol 15(4): e5219. DOI: 10.21769/BioProtoc.5219.
分类
发育生物学 > 形态建成 > 细胞结构
细胞生物学 > 细胞成像 > 共聚焦显微镜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link