- EN - English
- CN - 中文
Model Architecture Analysis and Implementation of TENET for Cell–Cell Interaction Network Reconstruction Using Spatial Transcriptomics Data
TENET模型架构分析及其在利用空间转录组学数据重建细胞间相互作用网络中的实现
(*contributed equally to this work, § Technical contact: yujianlee1119@gmail.com) 发布: 2025年02月05日第15卷第3期 DOI: 10.21769/BioProtoc.5205 浏览次数: 1431
评审: Prashanth N SuravajhalaAYŞE NUR PEKTAŞAnonymous reviewer(s)

相关实验方案
控制污染水平与深度测序 (CoLoC-seq) 相结合以探索细胞器转录组的全局定位拓扑
Anna Smirnova [...] Alexandre Smirnov
2023年09月20日 2690 阅读
Abstract
Cellular communication relies on the intricate interplay of signaling molecules, which come together to form the cell–cell interaction (CCI) network that orchestrates tissue behavior. Researchers have shown that shallow neural networks can effectively reconstruct the CCI from the abundant molecular data captured in spatial transcriptomics (ST). However, in scenarios characterized by sparse connections and excessive noise within the CCI, shallow networks are often susceptible to inaccuracies, leading to suboptimal reconstruction outcomes. To achieve a more comprehensive and precise CCI reconstruction, we propose a novel method called triple-enhancement-based graph neural network (TENET). The TENET framework has been implemented and evaluated on both real and synthetic ST datasets. This protocol primarily introduces our network architecture and its implementation.
Key features
• Cell–cell reconstruction network using ST data.
• To facilitate the implementation of a more holistic CCI, we incorporate diverse CCI modalities into consideration.
• To further enrich the input information, the downstream gene regulatory network (GRN) is also incorporated as an input to the network.
• The network architecture considers global and local cellular and genetic features rather than solely leveraging the graph neural network (GNN) to model such information.
Keywords: Cell–cell interaction network (CCI) reconstruction (细胞间相互作用网络(CCI)重建)Graphical overview
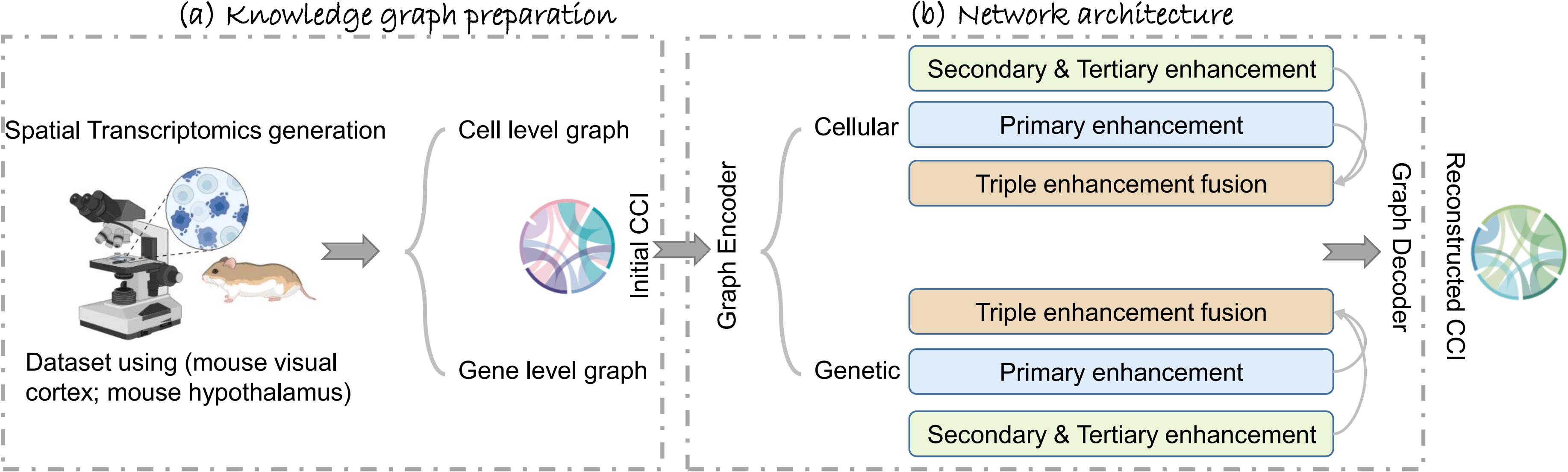
Graphical abstract of TENET, including (a) the knowledge graph preparation on both cell and gene levels and (b) the network architecture.
Background
Understanding cellular communication is crucial for constructing a cell–cell interaction network (CCI), which allows researchers to investigate the roles of different cells in biological processes and diseases. A common method for analyzing CCI involves studying the interactions between secreted ligands and their corresponding receptors (LR pairings), as these interactions are essential for signal transmission.
However, CCIs also occur through direct cell–cell contact, the extracellular matrix (ECM), and the secretion of signaling molecules [1]. Focusing solely on LR pairings overlooks the spatial context in which these interactions occur. With the advent of spatial transcriptomics (ST) data, incorporating spatial information can help address this limitation. Several methods for reconstructing CCI using ST data have been developed. For instance, Giotto [2] constructed a spatial grid based on cell coordinates to model proximal interactions but did not account for distal interactions. MISTy [3] utilizes multiple perspectives (intrinsic, local, tissue) to enhance cell knowledge for CCI inference. DeepLinc [4] employs a graph neural network (GNN) [5] to create a spatial proximity graph, using KNN [6] to identify both proximal and distal interactions. Despite these advances, modeling ST data solely at the cell level can lead to false positives and negatives due to a lack of downstream information. Incorporating downstream data, such as intracellular signaling, gene regulation, and protein changes can provide crucial insights for understanding actual CCI; CLARIFY [7] combines cellular and genetic information to build knowledge graphs for GNN inputs, effectively capturing structural features important for CCI inference. However, solely using GNNs is limiting, as they often aggregate local information, hindering the capture of global context. GNNs also struggle with similar nodes, which may lead to a loss of features’ specificity.
To overcome these challenges, we propose a novel approach called triple-enhancement-based graph neural network (TENET), designed as a comprehensive architecture for CCI reconstruction. TENET employs a graph convolution network (GCN) [8] backbone to extract global features from the data at multiple resolutions, generating robust latent feature embeddings. These embeddings then undergo a triple-enhancement mechanism that restores cell specificity that may have been lost during the GCN's global aggregation. The first enhancement mitigates the GCN's over-smoothing problem, while the secondary and tertiary enhancements refine the embeddings both locally and globally. Finally, a denoising loss function is introduced to tackle the challenges posed by noisy, low-quality data often encountered in small molecule experiments. This multi-scale enhancement strategy enables TENET to accurately and robustly reconstruct cell–cell interactions, addressing the limitations of previous methods and opening new avenues for researchers to explore the complexities of CCI.
Software and datasets
Software1. 2.90 GHz Intel i7-10700F CPU and NVIDIA A100 graphics card; nvcc: NVIDIA (R) Cuda compiler driver; Copyright 2005–2024 NVIDIA Corporation; Built on Tue_Feb_27_16:19:38_PST_2024; Cuda compilation tools, release 11.7, V11.7.99; Build cuda_11.7.r11.7/compiler.33961263_0
Input data
1. Three ST datasets are utilized, namely seqFISH, MERFISH, and scMultiSim [9], which can be acquired at https://bitbucket.org/qzhu/smfish-hmrf/src/master/, https://datadryad.org/stash/dataset/10.5061/dryad.8t8s248, and https://github.com/ZhangLabGT/scMultiSim. The hyperlinks direct to the datasets utilized in TENET. The specific analysis of the datasets is mentioned in the following sections.
Procedure
文章信息
稿件历史记录
提交日期: Jul 18, 2024
接收日期: Dec 25, 2024
在线发布日期: Jan 21, 2025
出版日期: Feb 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Wang, Z., Lee, Y., Xu, Y., Gao, P., Yu, C. and Chen, J. (2025). Model Architecture Analysis and Implementation of TENET for Cell–Cell Interaction Network Reconstruction Using Spatial Transcriptomics Data. Bio-protocol 15(3): e5205. DOI: 10.21769/BioProtoc.5205.
分类
生物信息学与计算生物学
系统生物学 > 空间转录组学
分子生物学 > RNA > RNA定位
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link