- EN - English
- CN - 中文
Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
一种简便高效的拟南芥胚胎RNA提取方法
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5194 浏览次数: 1911
评审: Noelia ForesiPooja SaxenaAnonymous reviewer(s)

相关实验方案

从标记的细胞类型中分离细胞核、提取RNA并去除核糖体RNA以用于RNA-Seq 分析
Mauricio A. Reynoso [...] Kaisa Kajala
2018年04月05日 16198 阅读
Abstract
Plant embryos are contained within seeds. Isolating them is crucial when endosperm and seed coat tissues interfere with the study of mutant genetic functions due to differing genotypes between maternal and embryonic tissues. RNA extraction from plant embryonic tissue presents particular challenges due to the high activity of RNases, the composition of the seed, and the risk of RNA degradation. The developmental stage of the embryo is a key aspect of successful isolation and RNA extraction due to the size and amount of tissue. Proper handling during RNA extraction is critical to maintain RNA integrity and prevent degradation. While commercial kits offer various methods for RNA extraction from embryos, homemade protocols provide valuable advantages, including cost-effectiveness and accessibility for labs with limited funding. Here, we present a simple and efficient protocol for extracting RNA from isolated Arabidopsis thaliana embryos at the torpedo/cotyledon stage using a homemade extraction buffer previously reported for styles of Nicotiana alata.
Key features
• This straightforward homemade protocol builds upon established methods for embryo isolation [1] and RNA extraction [2,3].
• It enables the extraction of high-quality RNA from Arabidopsis embryos at the torpedo stage.
• The protocol is designed to be both simple and cost-effective, making it an excellent option for labs with limited funding.
Keywords: Arabidopsis embryo (拟南芥胚胎)Graphical overview
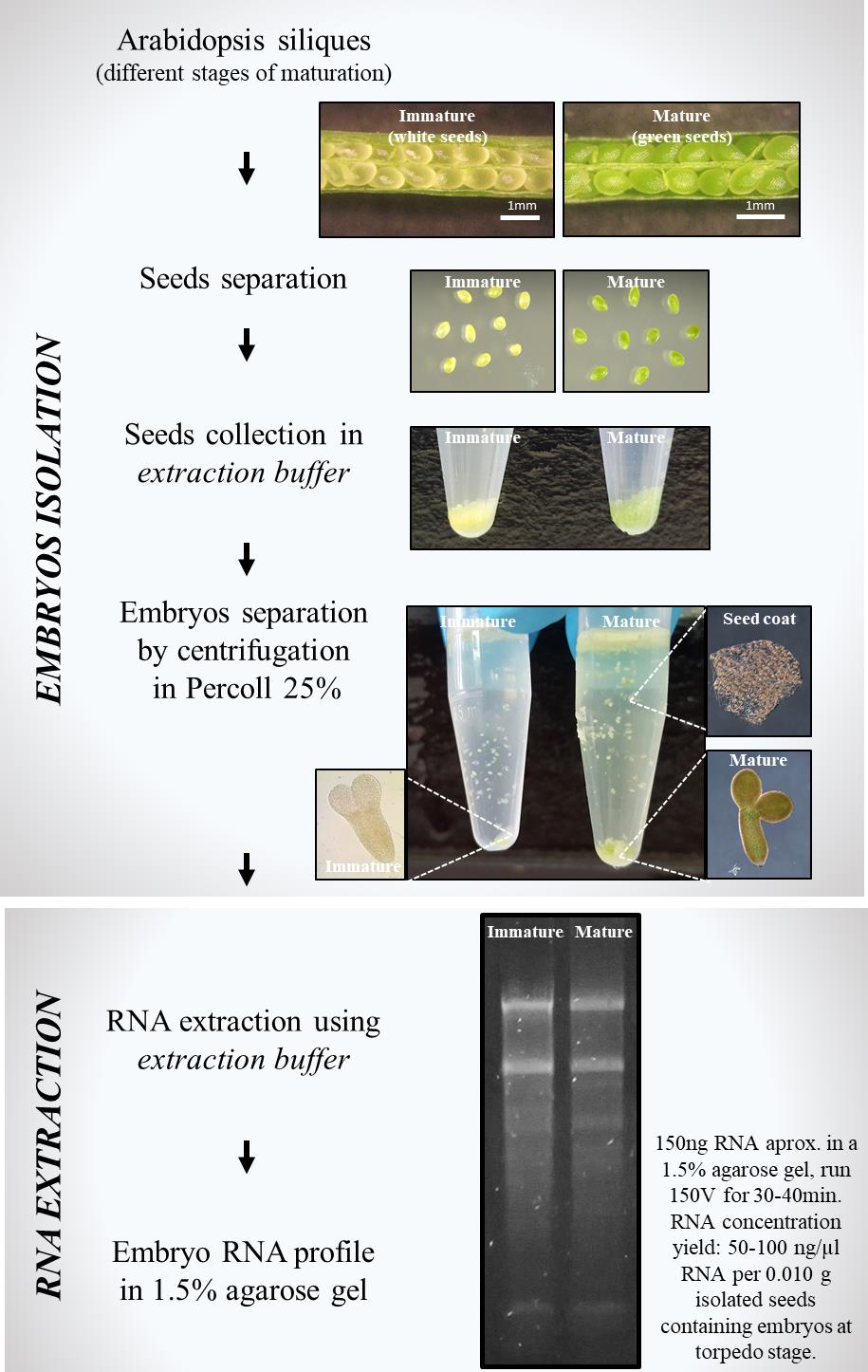
Simple method for RNA extraction from isolated Arabidopsis embryos using a homemade extraction buffer
Background
This protocol describes a straightforward two-step method for extracting RNA from isolated Arabidopsis embryos. Isolating RNA from plant tissues such as embryos, which are rich in RNases, poses significant challenges due to the risk of RNA degradation during sample preparation. To overcome these difficulties, commercial solutions such as RNAlater are commonly used to stabilize RNA by inactivating RNases, allowing for subsequent extraction using reagents like Trizol. In this protocol, we present a simple and effective homemade extraction buffer designed specifically for seed collection and RNA extraction. This buffer facilitates the isolation of high-quality RNA from Arabidopsis embryos. The method described here was successfully employed to extract RNA from developing embryos at the early torpedo and early cotyledon stages, from both immature (white) and mature (green) seeds. This approach offers an accessible and efficient alternative to commercial reagents, ensuring robust RNA recovery and preservation for downstream applications.
Materials and reagents
Biological materials
1. Arabidopsis thaliana ecotype Columbia (Col-0) wild type (WT) and embryo-lethal mutant plants from the Arabidopsis biological resource center (ARBRC, https://abrc.osu.edu/)
Reagents
1. Tris [Tri(hydroxymethyl)aminomethane] (Genbiotech, catalog number: RU2510)
2. EDTA (ethylenediamine tetraacetic acid disodium salt) (Biopack, catalog number: 1604.07)
3. Urea (Sigma, catalog number: U5378)
4. Sodium dodecyl sulfate (SDS) (Anedra, catalog number: 7211)
5. Ammonium acetate (Fluka, catalog number: 09690)
6. Lithium chloride (LiCl) (Sigma, catalog number: L9650)
7. 2-Mercaptoethanol pure (Biopack, catalog number: 9545.05)
8. Diethyl pyrocarbonate (DEPC) (Sigma, catalog number: D5758)
9. Percoll (GE Healthcare, catalog number: 17-0891-01)
10. DEPC (diethylpyrocarbonate) (Sigma, catalog number: D5758)
11. SYBRTM Safe DNA gel stain (Invitrogen, catalog number: S33102)
12. Agarose (Gene Biotech LE-Agarose 1200, catalog number: RU1010)
13. DEPC water (1 mL DEPC/L water; stirred overnight and autoclaved)
14. Phenol:chloroform:isoamyl alcohol 25:25:1 (Sigma, catalog number: P3803)
15. Chloroform (Biopack, catalog number: 1651.08)
16. Isopropanol (Mallinckrodt, catalog number: 3032-08)
Solutions
1. Buffer Tris HCl pH 8, 1 M (see Recipes)
2. EDTA pH 8, 0.5 M (see Recipes)
3. SDS 10% (see Recipes)
4. Ammonium acetate, 10 M (see Recipes)
5. Lithium chloride, 8 M (see Recipes)
6. Extraction buffer (see Recipes)
Recipes
1. Buffer Tris-HCl pH 8, 1 M
Weigh 12.11 g of Tris, add 75 mL of pure sterile water, adjust to pH 8 with HCl, and fill with pure sterile water to a final volume of 100 mL.
2. EDTA pH 8, 0.5 M
Weigh 18.6 g of EDTA, add 75 mL of pure sterile water, adjust to pH 8 using 10 M NaOH, and stir until EDTA is completely dissolved. Solubility improves while alkalinity increases; complete with pure sterile water to a 100 mL final volume.
3. SDS 10%
Dissolve 10 g of SDS in 100 mL of pure sterile water.
4. Ammonium acetate, 10 M
Dissolve 7.78 g of ammonium acetate in 10 mL of DEPC water.
5. Lithium chloride, 8 M
Dissolve 3.39 g of lithium chloride in 10 mL of DEPC water.
6. Extraction buffer (10 mL)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Urea | 7 M | 4.2 g |
| EDTA | 10 mM | 200 μL |
| Tris-HCl (1 M, pH 8) | 100 mM | 1 mL |
| SDS 10% | 1% | 1 mL |
| 2-Mercaptoethanol | 1% | 100 μL |
| H2O | to 10 mL | *see note |
*Note: Weigh urea in a 50 mL Falcon tube and dissolve urea in half volume (5 mL for a final volume of 10 mL) of water. Vortex the solution to help it dissolve. The bottom of smaller tubes hinders the full solubility of urea. Then, add the rest of the components and complete to final volume. Keep at room temperature. Cold will precipitate SDS in the extraction buffer. No risk for later RNA extraction.
Laboratory supplies
1. Syringe, needles (13 × 0.3 mm) and pointy tips tweezers (Dumont, catalog number: 0103-2-PO)
2. Plastic grinding rod for Eppendorf tubes (SSIbio, catalog number: 1005-39)
Equipment
1. Magnifying glass (LABKLASS, model: HG468405)
2. Table centrifuge (Eppendorf, model: centrifuge 5418)
3. Vortex (VornadoTM, Benchmark Scientific, catalog number/model: BV101-B)
4. Agarose gel electrophoresis equipment (Bio-Rad, model: PowerPacTM Basic Power Supply, Horizontal Electrophoresis Systems)
5. NanoDropTM One/One (Thermo Fisher, catalog number/model: ND-ONE-W)
Procedure
文章信息
稿件历史记录
提交日期: Sep 17, 2024
接收日期: Dec 10, 2024
在线发布日期: Jan 5, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Marchetti, F., Pagnussat, G. C. and Zabaleta, E. J. (2025). Simple Method for Efficient RNA Extraction From Arabidopsis Embryos. Bio-protocol 15(4): e5194. DOI: 10.21769/BioProtoc.5194.
分类
植物科学 > 植物分子生物学 > RNA > RNA 提取
分子生物学 > RNA > RNA 提取
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link