- EN - English
- CN - 中文
Campylobacter jejuni Biofilm Assessment by NanoLuc Luciferase Assay
NanoLuc荧光素酶法评估空肠弯曲菌生物膜
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5192 浏览次数: 1628
评审: Emilia KrypotouAnonymous reviewer(s)
Abstract
Campylobacter jejuni, a widespread pathogen found in birds and mammals, poses a significant risk for zoonosis worldwide despite its susceptibility to environmental and food-processing stressors. One of its main survival mechanisms is the formation of biofilms that can withstand various food-processing stressors, which is why efficient methods for assessing biofilms are crucial. Existing methods, including the classical culture-based plate counting method, biomass-staining methods (e.g., crystal violet and safranin), DNA-staining methods, those that use metabolic substrates to detect live bacteria (e.g., tetrazolium salts and resazurin), immunofluorescence with flow cytometry or fluorescence microscopy, and PCR-based methods for quantification of bacterial DNA, are diverse but often lack specificity, sensitivity, and suitability. In response to these limitations, we propose an innovative approach using NanoLuc as a reporter protein. The established protocol involves growing biofilms in microtiter plates, washing unattached cells, adding Nano-Glo luciferase substrate, and measuring bioluminescence. The bacterial concentrations in the biofilms are calculated by linear regression based on the calibration curve generated with known cell concentrations. The NanoLuc protein offers a number of advantages, such as its small size, temperature stability, and highly efficient bioluminescence, enabling rapid, non-invasive, and comprehensive assessment of biofilms together with quantification of a wide range of cell states. Although this method is limited to laboratory use due to the involvement of genetically modified organisms, it provides valuable insights into C. jejuni biofilm dynamics that could indirectly help in the development of improved food safety measures.
Key features
• Quantification of C. jejuni using NanoLuc luciferase.
• The assay is linear in the range of 1.9 × 107 to 1.5 × 108 CFU/mL.
• Following biofilm growth, less than 1 h is required for detection.
• The assay requires genetically modified bacterial strains.
Keywords: Campylobacter jejuni (空肠弯曲菌)Graphical overview
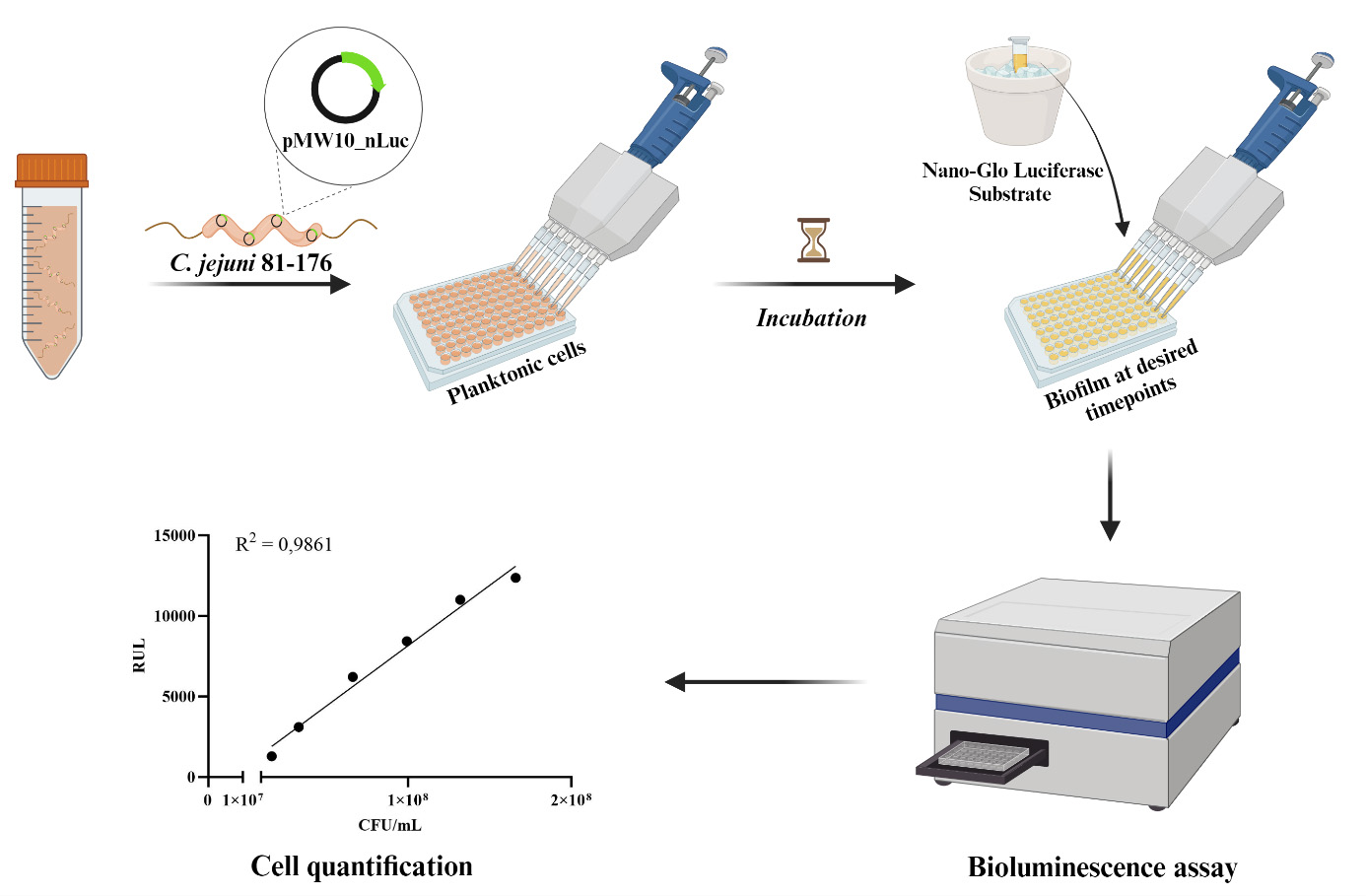
Campylobacter jejuni biofilm detection using NanoLuc luciferase assay
Background
Campylobacter jejuni, a pathogen found in the gastrointestinal tract of both domestic and wild birds and mammals, is a major cause of zoonosis worldwide [1]. However, it is sensitive to environmental stressors outside its hosts. To this end, C. jejuni forms biofilms as a survival strategy. Biofilms are characterized by a unique bacterial growth state and act as a protective shield that enables bacterial cells to resist certain environmental stressors—such as temperature fluctuations, desiccation, pH changes, and oxidative stress—commonly found on food preparation surfaces, food products, and water reservoirs [2–5]. Bacteria adapt to environmental stressors by, for example, adopting the nonmotile coccoid form and entering a viable but nonculturable state (VBNC) [6–8]. Viable but nonculturable cells cannot divide in vitro but maintain their membrane integrity and metabolic activity [9], which hinders their detection in quality control tests (a critical aspect of food safety) and may explain the persistence of campylobacteriosis in developed countries despite strict hygiene standards [1]. Studies suggest that bacteria in the VBNC state not only remain viable but also have the potential to recover and cause infections [10,11]. To assess biofilms, conventional cultivation methods and colony counts are generally used and usually complemented with biomass estimation using crystal violet staining [12,13]. The metabolic activity of biofilms can be assessed colorimetrically by monitoring the conversion of tetrazolium salt to formazan [14]. Simultaneously, ATP can be measured using BacTiter-Glo or resazurin fluorescence [8,12,13]. Advanced techniques such as transmission electron microscopy, scanning electron microscopy, and immunofluorescence with flow cytometry or fluorescence confocal laser scanning microscopy provide detailed information on the heterogeneity, cell localization, and structure of biofilms [15–17]. Additionally, PCR-based methods are used [12]. However, most of these methods are expensive, unspecific, technically demanding, and unsuitable for cell quantification. Therefore, new methods that can efficiently and rapidly detect C. jejuni cells would be valuable for advancing research related to food safety and possible applications in the food industry. The reporter protein NanoLuc offers several advantages, such as small size (19 kDa), high temperature stability (Tm = 60 °C), activity over a broad pH range (6–8), and the absence of post-translational modifications or disulfide bonds, which ensures uniform distribution within cells. Its most outstanding feature is its highly efficient bioluminescence. The reaction involves the NanoLuc enzyme, which is constitutively expressed, and the Nano-Glo substrate, commonly referred to as luciferin. Upon addition of the substrate in the presence of oxygen, NanoLuc (as luciferase) catalyzes the oxidation of luciferin and generates oxyluciferin. This molecule releases light in its excited energy state when it returns to its ground state [18,19]. No ATP is required for the luminescence reaction, which helps to minimize background luminescence. This reduction in the background signal ensures that the luminescence signal more accurately reflects the metabolic activity of the cells without external interference [20]. NanoLuc has already been successfully used to monitor and quantify Listeria innocua biofilms [21], and we have adapted the protocol to assess C. jejuni growth and biofilm formation. The protocol provides a comprehensive approach to biofilm monitoring and can be used as an important complement or alternative to conventional biofilm monitoring methods.
Materials and reagents
Biological materials
1. C. jejuni 81-176 (GenBank accession number NC_008787.1) transformed with pMW10_nLuc (GenBank accession number OR958835) (Figure 1)
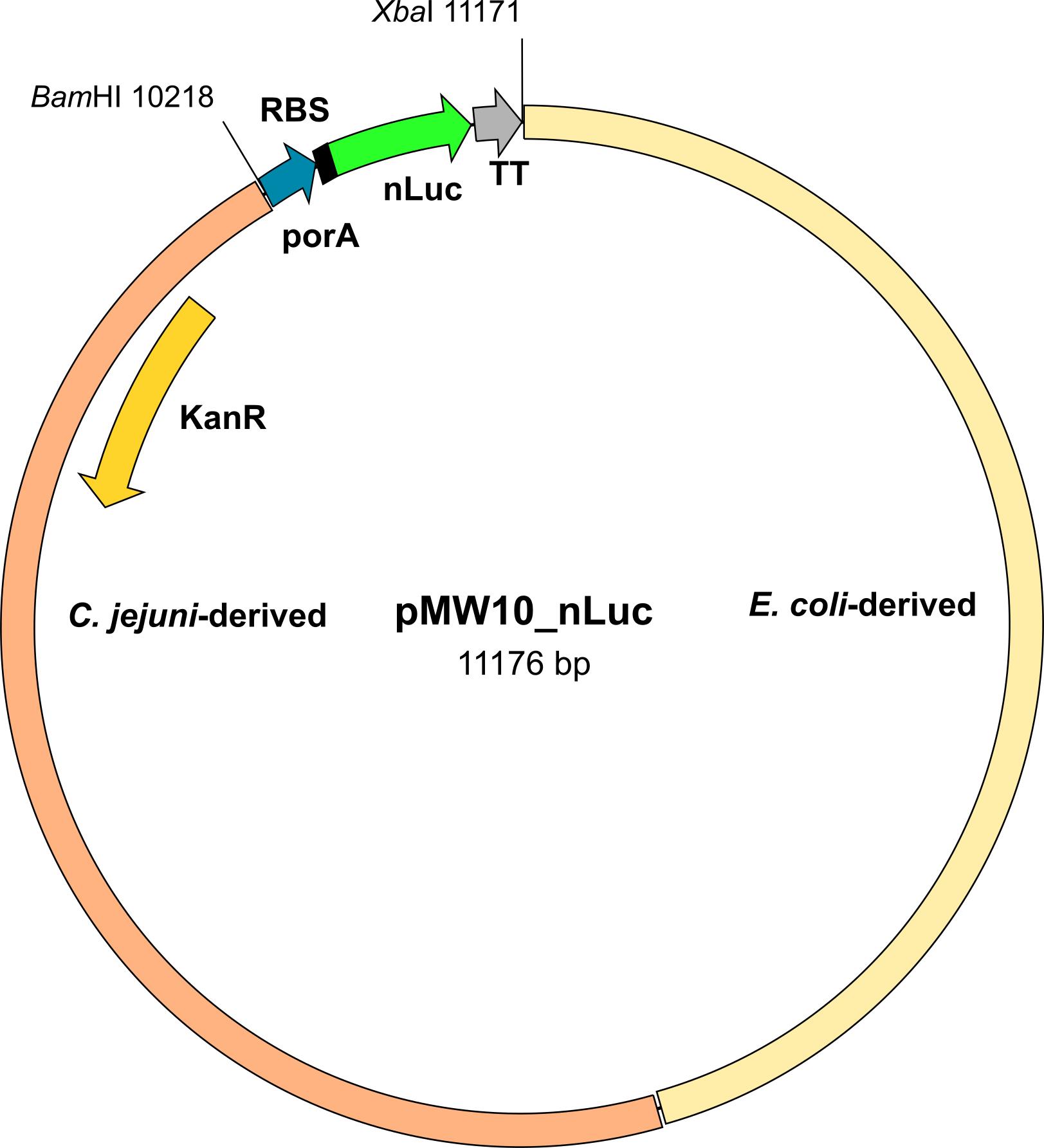
Figure 1. Scheme of the pMW10_nLuc plasmid. KanR, kanamycin resistance gene; porA, promotor; RBS, ribosome-binding site; nLuc, NanoLuc gene; TT, transcription terminator; BamHI and XbaI, restriction sites.
Reagents
1. Nano-Glo Luciferase Assay System (Promega, catalog number: N1120)
2. Phosphate-buffered saline (PBS) (Sigma-Aldrich, catalog number: P3813)
3. Mueller-Hinton broth (MHB) powder (BD Difco, catalog number: 275730)
4. Mueller-Hinton agar (MHA) powder (BD Difco, catalog number: 225250)
5. Karmali Campylobacter selective supplement (Oxoid, catalog number: SR0167E)
6. Ethanol, 99.8%, for HPLC, absolute (Thermo Scientific, catalog number: 445740010)
7. Kanamycin sulfate from Streptomyces kanamyceticus (BioReagent, Sigma-Aldrich, K1377)
Solutions
1. PBS solution (see Recipes)
2. MHB medium/MHA agar (see Recipes)
3. MHA Karmali agar (see Recipes)
Recipes
1. PBS solution
One pouch of PBS
1 L of purified water
Mix thoroughly. Autoclave at 121 °C for 15 min. Avoid overheating.
2. MHB medium/MHA agar
38 g of MHB powder or 21 g of MHA powder
1 L of purified water
Mix thoroughly. Heat with frequent agitation and boil for 1 min to completely dissolve the MHB or MHA powder. Autoclave at 121 °C for 15 min. Avoid overheating. Pour the MHA agar into sterile Petri dishes.
3. MHA Karmali agar
19 g of MHA powder
500 mL of purified water
One vial of Karmali Campylobacter selective supplement
1 mL of sterile distilled water
1 mL of 99.8% ethanol
Aseptically add 2 mL ethanol/sterile distilled water in a 1:1 ratio to the vial containing the Karmali Campylobacter selective supplement and mix gently to ensure that the contents dissolve completely. Add to 500 mL of the prepared MHA agar cooled to 50 °C (see Recipe 2). Mix well and pour into sterile Petri dishes.
Laboratory supplies
1. White flat-bottomed 96-well plates (Thermo Fisher, Microlite, catalog number: 7417)
2. 50 mL centrifuge tube (Corning, Mini Bioreactor, catalog number: 431720)
3. 1.5 mL microcentrifuge tubes (Eppendorf, Safe-Lock, catalog number: EP0030123611)
4. Pre-sterile 50 mL disposable reservoirs (Biotix, catalog number: EP0030123611)
5. Microplate sealing tape (Corning, catalog number: 3345)
6. Pipette tips 0.1–10 μL (Eppendorf, catalog number: 0030000811), 2–200 μL (Eppendorf, catalog number: 0030000889), 50–1,000 μL (Eppendorf, catalog number: 0030000927)
7. Inoculation loops (Golias, catalog number: EZ08)
Equipment
1. Incubator (Thermo Scientific)
2. Orbital shaker (Thermo Scientific)
3. Luminescence plate reader (BMG LabTech, FLUOstar Omega)
4. Pipette set (Eppendorf, catalog number: 05-403-151)
5. Multichannel pipette (Eppendorf, model: Research Plus, catalog number: 2231300048)
6. Biosafety cabinet (Nuaire)
7. Deep freezer (Haier, model: TwinCool)
8. Spectrophotometer (Bio-Rad, model: SmartSpec 3000, catalog number: 4006168)
9. Centrifuge (Thermo Scientific, model: LEGEND X1R, catalog number: 75004261)
Procedure
文章信息
稿件历史记录
提交日期: Sep 23, 2024
接收日期: Dec 12, 2024
在线发布日期: Jan 8, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Čukajne, T., Štravs, P., Sahin, O., Zhang, Q., Berlec, A. and Klančnik, A. (2025). Campylobacter jejuni Biofilm Assessment by NanoLuc Luciferase Assay. Bio-protocol 15(4): e5192. DOI: 10.21769/BioProtoc.5192.
分类
微生物学 > 微生物生物膜
微生物学 > 病原体检测 > 生物发光
环境生物学 > 细菌
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link