- EN - English
- CN - 中文
Precise Generation of Human Induced Pluripotent Stem Cell–Derived Cell Lines Harboring Disease-relevant Single Nucleotide Variants Using a Prime Editing System
使用Prime编辑系统精确生成携带与疾病相关的单核苷酸变异的来源于人诱导多能干细胞的细胞系
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5191 浏览次数: 2920
Abstract
Human induced pluripotent stem (iPS) cell lines harboring mutations in disease-related genes serve as invaluable in vitro models for unraveling disease mechanisms and accelerating drug discovery efforts. Introducing mutations into iPS cells using traditional gene editing approaches based on the CRISPR-Cas9 endonuclease often encounters challenges such as unintended insertions/deletions (indels) and off-target effects. To address these limitations, we present a streamlined protocol for introducing highly accurate gene mutations into human iPS cells using prime editing, a “search-and-replace” genome-editing technology that combines unwanted indel-minimized CRISPR-Cas9 nickase with reverse transcriptase. This protocol encompasses the design of prime editing guide RNAs (pegRNAs) required for binding and replacement at target loci, construction of prime editor and pegRNA expression vectors, gene transfer into iPS cells, and cell line selection. This protocol allows for the efficient establishment of disease-associated gene variants within 6–8 weeks while preserving critical genomic context.
Key features
• Dramatic improvement in efficiency of In-Fusion cloning using inserts assembled from the three pegRNA components (spacer, spCas9 scaffold, and 3' extension) via overlap extension PCR.
• Cost-effective and time-saving selection of pegRNAs for prime editing via bulk Sanger sequencing.
• Straightforward gene transfection using polymer-based reagents, which requires no specialized equipment or techniques and offers high reproducibility and broad applicability across different cell lines.
• Precise genome editing based on pegRNA/prime editing minimizes off-target effects, enabling a wide range of applications in the study of disease-associated genetic variants.
Keywords: Human induced pluripotent stem (iPS) cell (人诱导多能干细胞(iPS细胞))Graphical overview
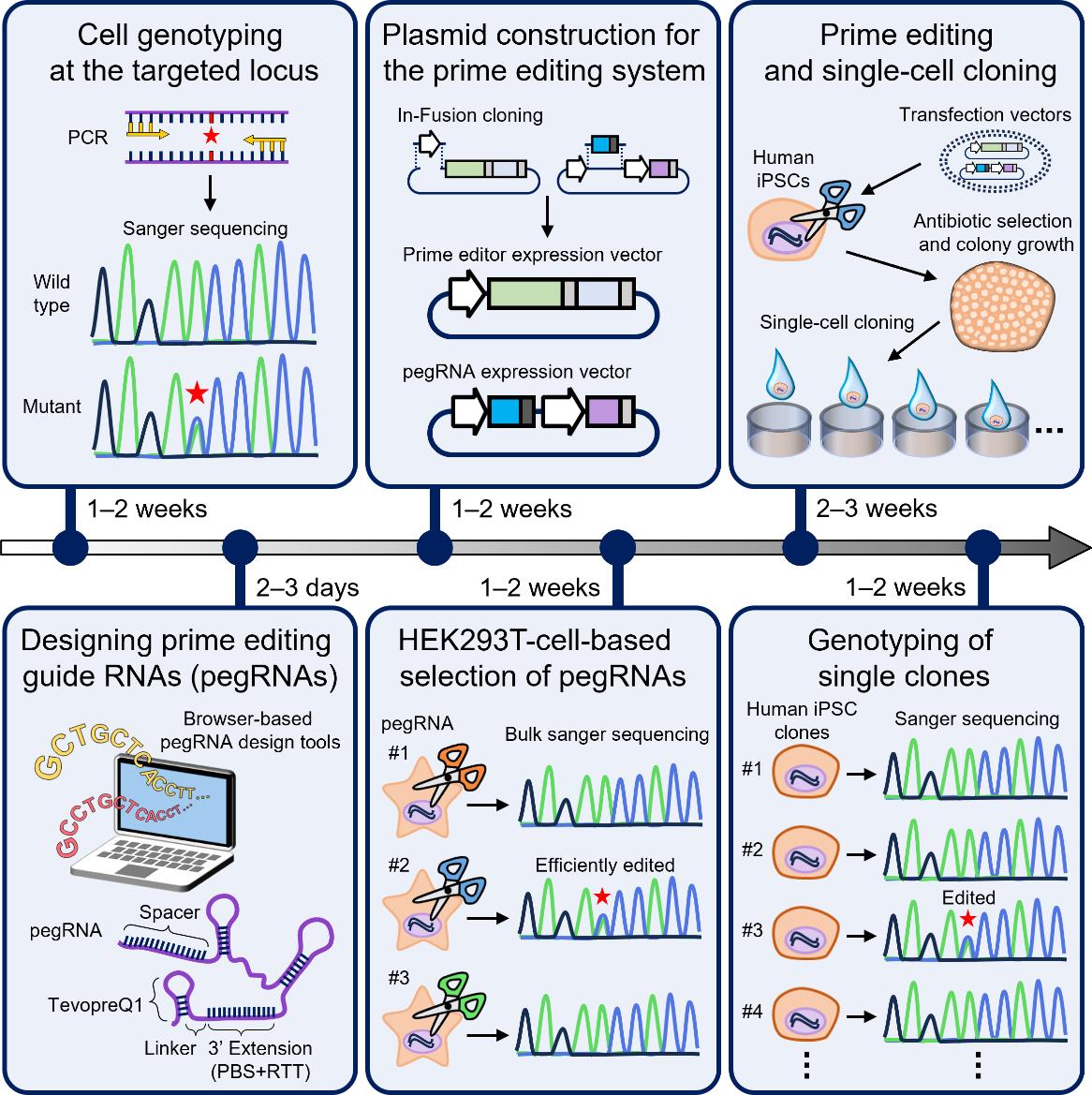
Key steps of generation of human induced pluripotent stem (iPS) cell lines harboring disease-relevant single nucleotide variants (SNVs) using a prime editing system
Background
Advances in genomic disease analysis have identified numerous single nucleotide variants (SNVs) associated with diverse diseases. Patient-derived human induced pluripotent stem (iPS) cells carrying these SNVs offer valuable in vitro models for elucidating disease mechanisms and accelerating drug discovery. However, phenotypic variability arising from diverse genetic backgrounds among iPS cell lines can confound the accurate assessment of disease gene effects [1].
Recent advancements in genome editing technologies have enabled the establishment of highly improved protocols for generating isogenic iPS cell lines through homology-directed repair (HDR)-mediated SNVs editing using CRISPR-Cas9 and single-stranded oligodeoxynucleotides (ssODNs), facilitating functional analyses of SNVs [2–5]. Nonetheless, these methods often introduce unintended insertions or deletions (indels) and off-target effects due to the induction of double-strand breaks in the DNA by CRISPR-Cas9, limiting their precision and efficiency [6].
To address these limitations, we present a protocol for generating iPS cell lines with precisely engineered SNVs in genes of interest using prime editing. Prime editing is a “search-and-replace” genome editing tool that combines a modified CRISPR-Cas9 nickase with reverse transcriptase to minimize unwanted indels [6]. This approach enables the functional analysis of gene variants while preserving the genomic context, providing a more controlled and informative system for studying disease mechanisms. Specifically, this protocol presents a workflow for generating a cell line harboring the OPTN p.(Asn51Thr) missense mutation, which was previously identified in our laboratory as an SNV associated with normal-tension glaucoma (NTG) [7].
This protocol has several key steps. First, the necessary components for the prime editing system are prepared, including the prime editor enzyme and prime editing guide RNA (pegRNA) expression vectors designed to introduce specific mutations. Additionally, a selectable marker gene, such as the puromycin resistance gene, is included to identify and select cells that have successfully taken up the editing components. Next, each pegRNA is introduced into HEK293T cells along with the prime editor enzyme. To assess the efficiency of the designed pegRNAs, the sequence of the target loci is analyzed in a bulk population of transfected cells. Finally, the pegRNA and prime editor enzyme are introduced into a human iPS cell line, 201B7, and individual iPS cell clones are isolated from the transfected population. By focusing on pegRNAs with high editing efficiency, the chances of obtaining iPS cell lines with the desired gene mutations are increased.
This protocol offers a robust and efficient method for generating iPS cell lines with targeted gene mutations, not only for studying disease-related genes but also for correcting pathogenic variants in patient-derived cells. While prime editing offers significant advantages over traditional methods, it is not without its limitations. For example, although several efficient pegRNA design tools are available, more information is needed in the future regarding editing efficiency in human iPS cells, as it can vary depending on the target locus and cell type. Furthermore, experimental conditions, such as reagent concentrations, transfection methods, and selection markers, must be carefully optimized for each iPS cell line to achieve optimal results.
Materials and reagents
Biological materials
1. Competent quick DH5α (Toyobo, catalog number: DNA-913F); store in a -80 °C ultra-low temperature freezer
2. Human iPS cell line 201B7 (RIKEN BRC Cell Bank, catalog number: HPS0063); to prevent a decrease in cell viability, iPS cells should be immediately transferred to liquid nitrogen for storage after receiving them from the supplier or after creating a cell bank
3. pLV[Exp]-EF1A>hCas9(ns):T2A:Puro (VectorBuilder, catalog number: VB210412-1054sbc)
4. pCMV-PEmax-P2A-hMLH1dn (Addgene, catalog number: 174828)
5. pU6-pegRNA-GG-acceptor (Addgene, catalog number: 132777)
6. pMK232 (CMV-OsTIR1-PURO) (Addgene, catalog number: 72834)
Store plasmids in a freezer at -30 °C.
Reagents
1. StemFit medium (Ajinomoto Healthy Supply, catalog number: AK03N)
2. DMEM (4.5 g/L glucose) with L-Gln and sodium pyruvate (Nacalai Tesque, catalog number: 08458-45)
3. Fetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: 12483020)
4. DPBS, no calcium, no magnesium (Thermo Fisher Scientific, catalog number: 14190144)
5. Trypsin-EDTA (0.25%), phenol red (Thermo Fisher Scientific, catalog number: 25200056); dilute with DPBS as needed.
6. TrypLE Select enzyme (1×), no phenol red (Thermo Fisher Scientific, catalog number: 12563011)
7. Penicillin-streptomycin (Thermo Fisher Scientific, catalog number: 15140122)
8. CultureSure Y-27632 (Fujifilm Wako Pure Chemical, catalog number: 036-24023)
9. iMatrix-511 (Matrixome, catalog number: 892012)
10. STEM-CELLBANKER GMP grade (Zenogen Pharma, catalog number: 11924)
11. PolyJet DNA in vitro transfection reagent (SignaGen Laboratories, catalog number: SL100688)
12. Puromycin (InvivoGen, catalog number: ant-pr)
13. QIAamp DNA Mini kit (Qiagen, catalog number: 51306)
14. NucleoSpin gel and PCR clean-up (Macherey-Nagel, catalog number: 740609.250)
15. SOC medium (Thermo Fisher Scientific, catalog number: 15544034)
16. Plasmid Miniprep Plus Purification kit (BioElegen Technology, catalog number: DP01-PLUS-300)
17. KOD One PCR Master Mix (Toyobo, catalog number: KMM-101)
18. Dpn I (New England Biolabs, catalog number: R0176L)
19. In-fusion snap assembly master mix (Takara Bio, catalog number: 638947)
20. Ampicillin sodium (Fujifilm Wako Pure Chemical, catalog number: 016-23301)
21. BD Difco LB (Luria-Bertani) broth Miller (Becton, Dickinson and Company, catalog number: 244620)
22. BD BACTO agar (Becton, Dickinson and Company, catalog number: 214010)
23. BD Difco 2×YT (yeast extract tryptone medium) (Becton, Dickinson and Company, catalog number: 244020)
24. Agarose S (Nippon Gene, catalog number: 318-01195)
25. Gel loading dye, purple (6×), no SDS (New England Biolabs, catalog number: B7025S)
26. 1 kb DNA ladder (Nippon Genetics, catalog number: NE-MWD1P)
27. 100 bp DNA ladder (Takara Bio, catalog number: 3422D)
28. Primers (OPC cartridge purified from Eurofins Genomics)
Homologous sequences for overlap PCR or in-fusion cloning are displayed in lowercase:
OPTN_genotyping_forward: 5'- AATCGCCAATGGGTTTGTGGGAC -3'
OPTN_genotyping_reverse: 5'- TGCTAAATCCTGTGCTTCCCCACC -3'
EF1A_ forward: 5'- ccagatatacgcgttGGCTCCGGTGCCCGTCAGTG -3'
EF1A_reverse: 5'- acggttcactaaaccTCACGACACCTGAAATG -3'
Inverse_PEmax_forward: 5'- ggtttagtgaaccgtCAGATCCGC -3'
Inverse_PEmax_reverse: 5'- aacgcgtatatctggCCCGTACATC -3'
EF1A_sequence_forward1: 5'- GGTGGAGACTGAAGTTAGGCCAGC -3'
EF1A_sequence_forward2: 5'- TAAGTGCAGTAGTCGCCGTGAACG -3'
EF1A_sequence_reverse: 5'- CACGCAAGGGCCATAACCCG -3'
tevopreQ1 + Terminator_forward: 5'- CGCGGTTCTATCTAGTTACGCGTTAAACCAACTAGAA -3'
tevopreQ1 + Terminator_reverse: 5'- AAAAAATTCTAGTTGGTTTAACGCGTAACTAGATAGAACCGCG -3'
Spacer_OPTN_forward: 5'- gaaaggacgaaacaccGCTGCTCACCTTTCAGCTGG -3'
Spacer_OPTN_reverse: 5'- ttctagctctaaaacCCAGCTGAAAGGTGAGCAGC -3'
spCas9_scaffold_forward: 5'- GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGA -3'
spCas9_scaffold_reverse: 5'- GCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGC -3'
3’extension + Linker_OPTN_forward: 5'- gcaccgagtcggtgcTGACCGAGACCCACCAGCTGAAAGGACCC -3'
3’extension + Linker_OPTN_reverse: 5'- ctagatagaaccgcgATTAGGGTCCTTTCAGCTGGTGGGTCTCGGTC -3'
Inverse_U6-pegRNA_Forward: 5'- cgcggttctatctagTTACGCG -3'
Inverse_U6-pegRNA_reverse: 5'- ggtgtttcgtcctttcCACAAGA -3'
pegRNA_sequence_forward: 5'- GAGGGCCTATTTCCCATGATTCC -3'
Laboratory supplies
1. 15 mL centrifuge tubes (TPP, catalog number: 91015)
2. 1.5 mL microcentrifuge tubes (Ina-optika, catalog number: 113004)
3. 0.5 mL microcentrifuge tube (Ina-optika, catalog number: SC-005)
4. 20 μL racked tips, sterile (Eppendorf, catalog number: 0030075226)
5. 200 μL racked tips, sterile (Eppendorf, catalog number: 0030075234)
6. 1000 μL racked tips, sterile (BM Equipment, catalog number: WE1000-RL)
7. 12-well cell culture plate (Corning, catalog number: 353043)
8. 24-well cell culture plate (Corning, catalog number: 353047)
9. 48-well cell culture plate (Corning, catalog number: 353078)
10. 96-well cell culture plate (Corning, catalog number: 353072)
11. 60 × 15 mm cell culture dish (Corning, catalog number: 353004)
12. STAR SDish9015 ver. 2 Petri dish (Rikaken, catalog number: RSU-SD9015-2)
13. 0.2 mL 8-tube PCR strips (Bio-Rad Laboratories, catalog number: TLS0801)
14. 0.2 mL domed PCR tube 8-cap strips (Bio-Rad Laboratories, catalog number: TCS0801)
15. 2 mL cryo vial, inner screw (Simport Scientific, catalog number: T311-2)
16. BICELL freezing treatment container (Nihon Freezer, catalog number: BICELL)
Equipment
1. Thermal cycler (Bio-Rad, model: C1000 Touch)
2. Refrigerated centrifuge (Tomy Seiko, model: LX-120)
3. High-speed refrigerated microcentrifuge (Tomy Seiko, model: MX300)
4. Cell culture incubator (PHC, catalog number: MCO-170AICUVD-PJ)
5. Shaker incubator (Taitec, model: BR-40LF)
6. Benchtop incubator for bacterial culture (AS ONE, model: PIC-100)
7. Microvolume spectrophotometer (Thermo Fisher Scientific, model: NanoDrop 2000c)
8. Gel electrophoresis equipment (Takara Bio, model: Mupid-exU)
9. Gel imaging system (Bio-Rad, model: GelDoc Go imaging system)
10. Autoclave (Tomy Seiko, model: LBS-245)
Software and datasets
1. SnapGene Viewer (SnapGene, https://www.snapgene.com/snapgene-viewer)
Procedure
文章信息
稿件历史记录
提交日期: Oct 27, 2024
接收日期: Dec 12, 2024
在线发布日期: Jan 5, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Kanno, S., Sato, K. and Nakazawa, T. (2025). Precise Generation of Human Induced Pluripotent Stem Cell–Derived Cell Lines Harboring Disease-relevant Single Nucleotide Variants Using a Prime Editing System. Bio-protocol 15(4): e5191. DOI: 10.21769/BioProtoc.5191.
分类
干细胞 > 多能干细胞 > 基于细胞的分析方法
分子生物学 > DNA > 转染
医学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link