- EN - English
- CN - 中文
Novel Workflows for Separate Isolation of Pathogen RNA or DNA From Wastewater: Detection by Innovative and Conventional qPCR
废水中病原RNA或DNA的分离新型工作流程:创新与传统qPCR检测法
(*contributed equally to this work) 发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5189 浏览次数: 3193
评审: Lucy XieSonali ChaturvediWeidong An

相关实验方案

优化高分子量 DNA 提取方法以用于 Magnaporthaceae 及其他禾本科根部真菌的长读长全基因组测序
Michelle J. Grey [...] Mark McMullan
2025年03月20日 3249 阅读
Abstract
Wastewater-based surveillance (WBS) can provide a wealth of information regarding the health status of communities from measurements of nucleic acids found in wastewater. Processing workflows for WBS typically include sample collection, a primary concentration step, and lysis of the microbes to release nucleic acids, followed by nucleic acid purification and molecular-based quantification. This manuscript provides workflows from beginning to end with an emphasis on filtration-based concentration approaches coupled with specific lysis and nucleic acid extraction processes. Here, two WBS processing approaches are presented, one focusing on RNA-specific pathogens and the other focused on DNA-specific pathogens found within wastewater: 1) The RNA-specific approach, employed for analyzing RNA viruses like severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) couples electronegative filtration of wastewater with the placement of the filter within a lysis buffer followed by direct RNA extraction. 2) The DNA-specific approach, employed for analyzing DNA pathogens like Candida auris, uses size selection membranes during filtration, subsequently followed by a lysis buffer, bead-beating, and DNA extraction. Separate workflows for RNA versus DNA isolations have the advantage of improving the detection of the target pathogen. A novel aspect of the RNA-specific workflow is the direct extraction of nucleic acids from filter lysates, which shows enhanced recoveries, whereas the DNA-specific approach requires bead beating prior to extraction. Novelty is also provided in a new qPCR approach called Volcano 2nd Generation (V2G), which uses a polymerase capable of using RNA as a template, bypassing the reverse transcriptase step normally required for qPCR.
Key features
• Membrane filtration approaches for concentrating suspended solids from wastewater. After concentration, workflows are optimized for separate recovery of RNA and DNA.
• Unique polymerase utilized to perform qPCR analysis, foregoing reverse transcription, for RNA.
• Sample products for use with other molecular techniques (e.g., sequencing) as workflow approaches generate high-quality, concentrated nucleic acid extracts with minimal inhibitors.
• Validated through COVID-19 surveillance where >1,000 samples of wastewater and >3,000 filter concentrates produced from these samples have been created and analyzed, with published results.
Keywords: Wastewater (废水)Graphical overview
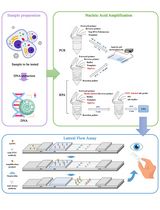
Laboratory workflow of sample preparation
Background
The use of wastewater for surveying the health of communities has been a growing application in the field of public health. Analyzing environmental samples like wastewater can inform researchers of current infectious disease spread or the general health of communities. More specifically, wastewater can capture spatial and temporal trends of pollutants, drug abuse in communities, and antimicrobial resistance genes [1]. Recent wastewater-based surveillance (WBS) literature has emphasized not only WBS’s rapid development but also shed light on the importance of method selection prior to experimentation. The paper by Pecson et al. [2] is a primary example of such literature, where 32 participating laboratories at the start of the COVID-19 pandemic cooperated to compare strategies for pathogen surveillance from wastewater. Their focus was on reproducibility, sensitivity, and the impact that “other method steps” have on the recovery of viruses like SARS-CoV-2. Since the publication of Pecson et al. [2], many researchers have initiated WBS programs and have focused on comparing specific methods; a general theme has been to expand WBS’s application for SARS-CoV-2 detection to other pathogens of interest to expand WBS as a tool for public health [3–11,12,13].
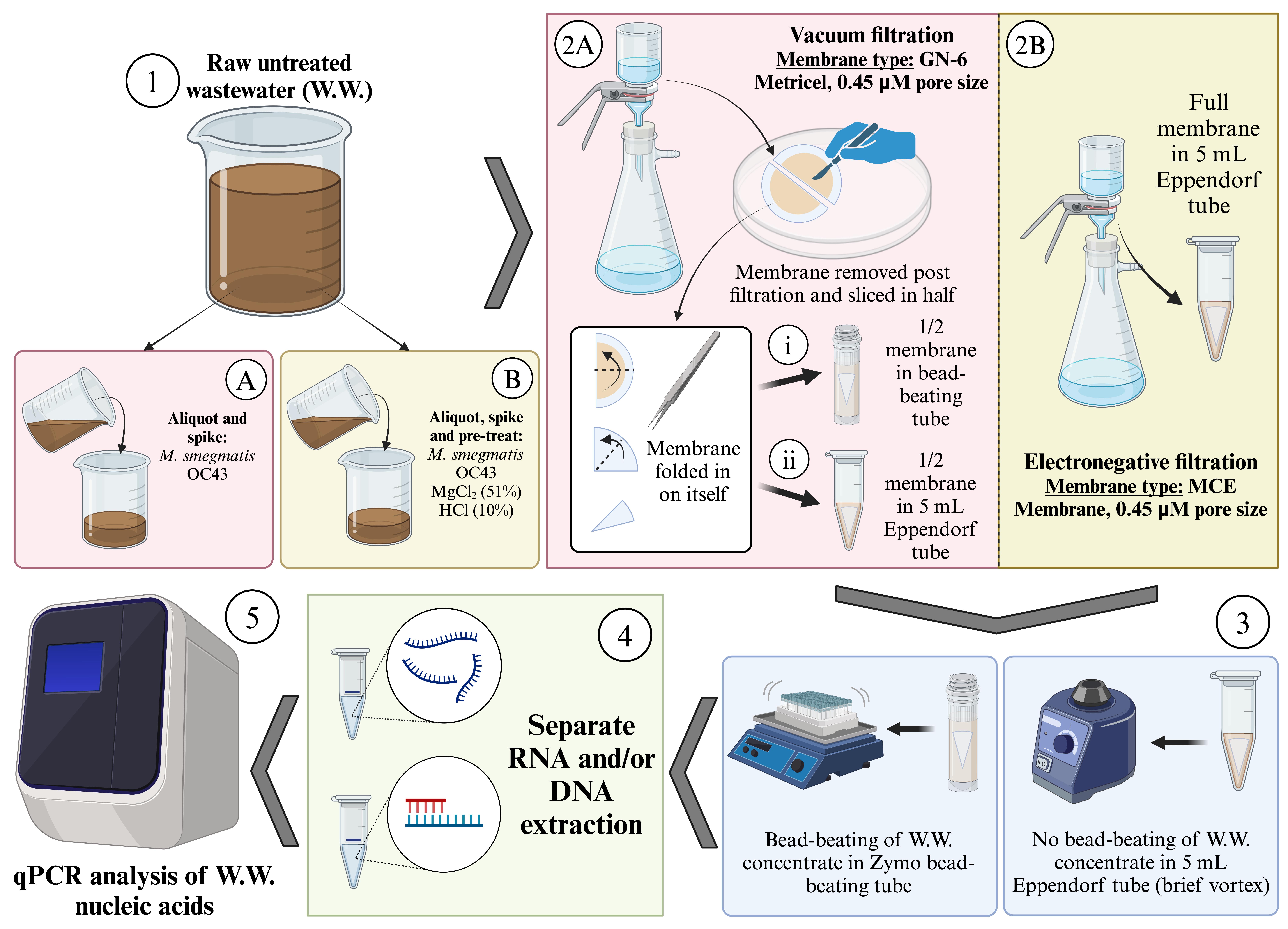
Although methods are merging toward the isolation of total nucleic acids (RNA and DNA simultaneously) using the same extraction procedure, studies have shown [2] that optimal recoveries require different extraction procedures. Here, we propose two different concentration-plus-extraction strategies—one for the recovery of RNA and another for the recovery of DNA, both optimized for wastewater. The assessment of samples with the Volcano 2nd Generation (V2G) qPCR assay adds an additional uniqueness to this procedure due to the equal performance of V2G to conventional RT-qPCR [6] while providing a decrease in sample analysis time. While filtration-based concentration is not a novel concept in and of itself, its use, coupled with sample type, membrane selection, filtration protocol, lysis method, and downstream molecular techniques, is unique and adjustable for specific research needs. For example, varying volumes of wastewater can be used depending on the composition of each sample (turbidity, suspended solids concentration, etc.). Additionally, different membrane types and pore sizes can be utilized to fit the specific application. The filtration approach can be optimized for the capture of viruses by charge or for larger microbes by size exclusion. This includes pathogens like Candida auris, a growing health concern in many communities [14–16]. The use of bead beating, especially coupled with filtration approaches, is also advantageous as it increases the likelihood of detecting pathogens with cell membranes like C. auris [16] from wastewater. However, bead beating is not recommended for the recovery of RNA viruses. Bead beating can limit the detection of RNA viruses due to heat and mechanical agitation, which can lead to the degradation of some nucleic acids if not optimized prior to experimentation [3]; it also extracts ribosomal RNA from bacteria and higher forms of microbes, confounding the RNA signal from viruses. The inclusion of chemicals like HCl for the recovery of viruses by charge, although recommended in this protocol, could also impact recovery, as seen in Babler et al. [3]. Regarding limitations, filtration-based workflows require sterilization of equipment unless disposable options are favored, so they can be costly to initiate. Moreover, the processes presented here from start to finish are designed to be completed manually as opposed to magnetic bead concentration using robotic approaches [18], so laboratory capacity and personnel bandwidth also play into the ability to use these methods if many samples are expected to be analyzed. To expand these protocols to evaluate pathogens beyond SARS-CoV-2 and C. auris, optimization is recommended to ensure that recoveries of the intended pathogen are suitable for downstream applications. These methods, as they have been optimized for wastewater, may not be generalizable for other sample types.
Materials and reagents
Reagents
Electronegative filtration and RNA extraction workflow: specific materials and reagents
1. Hydrochloric acid 10% (Spectrum Chemical MFG Corp., catalog number: HY105)
2. pH standards 4, 7, 10 (Cole-Parmer, catalog number: UX-05942-10)
3. Magnesium chloride, MgCl2, 51% w/v aqueous solution (RICCA Chemical Company, catalog number: 4470)
4. Zymo QuickRNA Viral Kit (Zymo Research, catalog number: R1034/1035)
5. V2G buffer (MyPols, catalog number: 8100)
6. V2G polymerase (MyPols, catalog number: 8400M)
7. Rox (Thermo Fisher Scientific, catalog number: 12223012)
8. dNTPs (Thermo Fisher Scientific, catalog number: R0193)
9. Platinum Taq Antibody (TaKaRa, catalog number: 9002A)
Vacuum filtration and DNA extraction workflow: specific materials and reagents
1. ZymoBIOMICS DNA Miniprep kit (Zymo Research, catalog number: D4300)
2. TaqMan Fast Universal PCR master mix (2×) (Thermo Fisher, catalog number: 4352042)
Materials and reagents used for both workflows
1. Vero Cells [American Type Culture Collection (ATCC), catalog number: CCL-81]
2. Luria Bertani (LB) Broth (Thermo Fisher Scientific, catalog number: 10855001)
3. LB agar plates (Thermo Fisher Scientific, catalog number: 22700025)
4. Tween 80 (Millipore Sigma, catalog number: P1754-25ML)
5. 100 mg/mL ampicillin (Millipore Sigma, catalog number: A5354-10ML)
6. Mycobacterium smegmatis mc2155 (NIH AIDS Reagent Program, catalog number: 2195)
7. Beta coronavirus 1, strain OC43 (ATCC, catalog number: VR-1558)
8. RPMI media (Thermo Fisher Scientific, catalog number: 12633012)
9. Wastewater [collected in pre-labeled pre-sterilized bottles (500 mL capacity) containing 0.5 mL of sterile 100 g/L sodium thiosulfate; details of bottles and reagents below]
10. 1× DNA/RNA shield (Zymo Research, catalog number: R1100-250)
11. OneStep PCR Inhibitor Removal kit (Zymo Research, catalog number: D6030)
12. qPCR primers (synthesized by Sigma-Aldrich)
13. qPCR probes (synthesized by Integrated DNA Technologies)
14. Nuclease-free water (Fisher Scientific, catalog number: AM9930)
15. 99.5% isopropanol (Thermo Fisher Scientific, catalog number: 149320050)
16. 70% ethanol (Fisher Scientific, catalog number: BP82031GAL)
17. 100% ethanol (Fisher Scientific, catalog number: BP2818500)
18. Sodium thiosulfate (GrowingLabs, catalog number: LC250001)
19. Clorox bleach (concentrated)
Laboratory supplies
Electronegative filtration and RNA extraction workflow: specific laboratory supplies
1. MCE membrane filter, 47 mm, 0.45 μm pore size (Millipore Sigma, catalog number: HAWP04700)
2. Magnetic stir bars [12 mm (L) × 3 mm (D)] (VWR, catalog number: 58948-091)
3. DI water (Thermo Fisher Scientific, catalog number: 15230001)
4. Glass dropper (Fisher Scientific, catalog number: 14-955-502)
5. 5 mL Eppendorf Tubes (sterile) (Eppendorf, catalog number: 0030119460)
Vacuum filtration and DNA extraction workflow: specific laboratory supplies
1. Pall GN-6 Metricel MCE membrane disc filters, 47 mm, 0.45 μm pore size (New Star Environmental, catalog number: 66278)
2. 2 mL ZR BashingBead lysis tubes (0.1 & 0.5 mm) (Zymo Research, catalog number: S6012-50)
3. Polystyrene, disposable, sterile Petri dishes (Carolina, catalog number: 741248)
4. Disposable sterile scalpels #10 (Surgical Supply Service, catalog number: 75745)
Laboratory supplies used for both workflows
1. Biohazard waste benchtop container (1.4 L) (VWR, catalog number: 11214-708)
2. Vactrap 2 vacuum trap system for aspiration and vacuum protection (VWR, catalog number: 76207-602)
3. Pall magnetic filter funnels, 47 mm [Weber Scientific, catalog number: EF8452B (300 mL), EF8452F (500 mL)]
4. Clamps and chains to hold sample collection bottles used to collect samples from sewer holes in the field [available from any hardware store. Hose clamps large enough to attached to bottles (MIAHART, 6-inch hose clamp adjustable 304 stainless steel duct clamps worm gear), 1/8-inch chain hooks with locking connector (BNYZWOT, D-shape, twist lock), and lightweight utility chain are recommended (4every SUS304). The chain hooks are used to connect the hose clamps holding the bottles to two chains, one on each side of the bottle]
5. Strainer/colander for field (Bradshaw Home, catalog number: 72115); the strainer should have 2–3 mm diameter openings, kitchen type strainer will work
6. Funnel for field (Walmart, catalog number: HTFF-2020); the funnel should be wide at the top and narrow enough at the bottom to fit easily into the sample collection bottles
7. 5-gallon heavy-duty plastic buckets to capture spillage when pouring wastewater samples (Lowe’s, catalog number: 954434)
8. 2 L sterile plastic wide-mouth collection bottles (Thermo Fisher Scientific, catalog number: 2120-0005 if disposable, or Thermo Fisher Scientific, catalog number: 2121-0005 for reuse via autoclaving)
9. 500 mL sterile plastic collection bottles (VWR, catalog number: 16060-012 if disposable, or VWR, catalog number: 16060-012 for reuse via autoclaving); a 500 mL volume line marked on the bottle using a paint pen
10. 1 L sterile wide-mouth collection bottles (VWR, catalog number: 16060-014 if disposable, or VWR, catalog number: 16060-014 for reuse via autoclaving)
11. Kimwipes (Grainger, catalog number: 36VC06)
12. Graduated cylinders made of polypropylene for reuse by autoclaving (100 mL) (Carolina, catalog number: 721603)
13. Spade smooth tip forceps (AmScope, catalog number: TW-487)
14. PYREX griffin beakers (100 mL) (Millipore Sigma, catalog number: CLS1000100-12EA)
15. P5000 pipette (Gilson, catalog number: F144066)
16. P5000 pipette tips (Thermo Fisher Scientific, catalog number: 94052550)
17. P1000 pipette (Gilson, catalog number: F144059M)
18. P1000 filtered pipette tips (IBIS Scientific, catalog number: M-1000-9FC)
19. P200 Pipette (Gilson, catalog number: F144058M)
20. P200 filtered pipette tips (IBIS Scientific, catalog number: M-0200-9FC)
21. P100 pipette (Gilson, catalog number: F144057M)
22. P100 filtered pipette tips (IBIS Scientific, catalog number: M-0100-9FC)
23. P20 pipette (Gilson, catalog number: F144056M)
24. P20 filtered pipette tips (IBIS Scientific, catalog number: M-0020-9FC)
25. P10 pipette (Gilson, catalog number: F144055M)
26. P10 pipette tips (IBIS Scientific, catalog number: M-0010-9FC)
27. Tube racks (Eppendorf, 1.5 mL and 2 mL, catalog number: 0030119819; 5 mL and 15 mL, catalog number: 0030119827)
28. Autoclave bins (Fisher Scientific, catalog number: 13-359-20B)
29. T175 cell culture flasks (Fisher Scientific, catalog number: 10-126-34)
30. Serological pipettor (Pipette.com, catalog number: DP-501R)
31. 10 mL serological pipettes (Fisher Scientific, catalog number: NC9868325)
32. 50 mL conical tubes (Fisher Scientific, catalog number: 14-432-22)
33. 96-well plates (Bio-Rad, catalog number: HSP9601)
34. Bio-Rad microseal “B” seal (Bio-Rad, catalog number: MSB1001)
35. 1.5 mL microcentrifuge tubes (Fisher Scientific, catalog number: 05-408-129)
36. Labels for all tubes (Brady, catalog number: THT-59-7425-2-SC)
37. Freezer storage boxes (81 slot: Alkali Scientific, catalog number: FB2CC-81; 25 slot: Eppendorf, catalog number: 0030140532)
38. Laboratory coats (Fisher Scientific, catalog number: 19-181-594)
39. Disposable laboratory coats for field (Dupont Tyvek 400, catalog number: TY212S)
40. Gloves (VWR, catalog number: 76582-340)
41. Eye protection (Fisher Scientific, catalog number: 19-039-590)
42. (Optional) respiratory protection (Breatheze KN95 disposable face masks, Amazon)
43. Laboratory data sheets on clipboard with pen (created depending on lab needs using Microsoft Word or Excel)
44. Permanent paint pen (ULINE, catalog number: S-20622)
Equipment
Electronegative filtration and RNA extraction workflow: specific equipment
1. pH Meter (Orion Star) (Thermo Fisher Scientific, catalog number: STARA2110)
2. Magnetic stir plate (GrowingLabs, catalog number: H4000-S)
3. Vortex Genie 2 (Scientific Industries, Inc., catalog number: SI-0236)
Vacuum filtration and DNA extraction workflow: specific equipment
1. BeadRuptor 12 Instrument (Omni International)
Equipment used for both workflows
1. YSI probe (Xylem YSI ProDSS, catalog number: 626870-1); for measuring basic water quality of samples. Recommend adding probes for pH (#626904), water temperature and salinity (#626902), turbidity (#626901), and dissolved oxygen (#626900)
2. Clean water reservoir with hand pump to rinse of equipment after sample collection (Itisll, portable garden pump sprayer, Size: 2 Gal_Brasswand, ULINE, catalog number: S-20860)
3. (Optional) Refrigerated autosampler (HACH, catalog number: AS950 fitted with an IO9000 for flow proportional sampling) or non-refrigerated autosampler (Teledyne ISCO, catalog number: ISCO 6712, fitted with a 2150 area velocity meter for flow paced sampling; otherwise, use time paced). Refrigerated with flow measurement preferred
4. BSL-2 hood
5. Whatman vacuum manifold (LabFilterz, catalog number: 10498761)
6. MyFuge centrifuge (5 mL capacity) (Grayline Medical, catalog number: C1005)
7. Eppendorf centrifuge (2 mL capacity) (Eppendorf, catalog number: 5420000245)
8. Benchtop centrifuge (Southern Labware, catalog number: C1008-B)
9. Bio-Rad CFX connect RealTime qPCR instrument (Bio-Rad Laboratories, catalog number: 1855201)
10. Plate spinner (GrowingLabs, catalog number: C2000-115V)
11. Synergy BioTek plate reader (Fisher Scientific, catalog number: BTS1LASI)
12. 37 °C shaker/incubator (Fisher Scientific, catalog number: 50-195-3922)
13. Spectrophotometer (Eppendorf, model: 6135)
14. Culture plate rocker (Millipore Sigma, catalog number: Z742529-1EA)
15. Corning LSE digital dry bath heater (Fisher Scientific, catalog number: 07-203-024)
16. Igloo or Coleman cooler (purchased from any general store)
17. Access to 4 °C, -20 °C, and -80 °C refrigerators and freezers. Freezers should not include defrost cycles to facilitate the preservation of samples in the long term
18. Access to autoclave for sterilizing reusable equipment
Software and datasets
1. BioRender (https://www.biorender.com/). The following figures were created using BioRender: Graphical overview, Babler, K. (2024) https://www.BioRender.com/q34x703
Procedure
文章信息
稿件历史记录
提交日期: Jul 5, 2024
接收日期: Dec 8, 2024
在线发布日期: Jan 14, 2025
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Babler, K. M., Solo-Gabriele, H. M., Sharkey, M. E. and Amirali, A. (2025). Novel Workflows for Separate Isolation of Pathogen RNA or DNA From Wastewater: Detection by Innovative and Conventional qPCR. Bio-protocol 15(4): e5189. DOI: 10.21769/BioProtoc.5189.
分类
环境生物学 > 生态系统
微生物学 > 病原体检测 > PCR
分子生物学 > DNA > DNA 提取
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link