- EN - English
- CN - 中文
Protocol to Mine Unknown Flanking DNA Using PER-PCR for Genome Walking
使用PER-PCR进行基因步移挖掘未知侧翼DNA的实验方案
(*contributed equally to this work) 发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5188 浏览次数: 1494
评审: Alba BlesaMichael KotikAnonymous reviewer(s)
Abstract
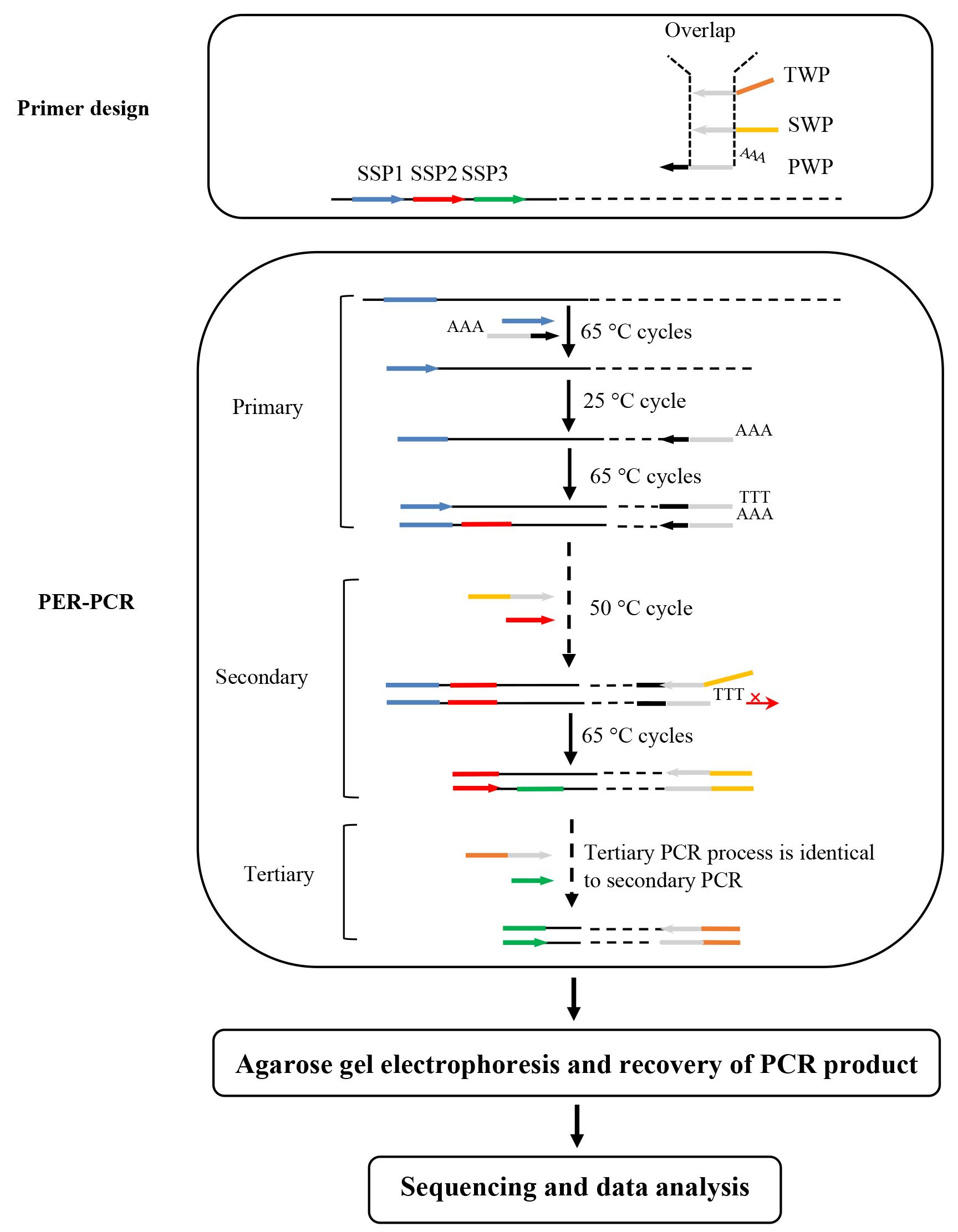
Genome walking, a molecular technique for mining unknown flanking DNAs, has a wide range of uses in life sciences and related areas. Herein, a simple but reliable genome walking protocol named primer extension refractory PCR (PER-PCR) is detailed. This PER-PCR-based protocol uses a set of three walking primers (WPs): primary WP (PWP), secondary WP (SWP), and tertiary WP (TWP). The 15 nt middle region of PWP overlaps the 3' region of SWP/TWP. The 5' regions of the three WPs are completely different from each other. In the low annealing temperature cycle of secondary or tertiary PER-PCR, the short overlap mediates the annealing of the WP to the previous WP site, thus producing a series of single-stranded DNAs (ssDNA). However, the 5' mismatch between the two WPs prevents the template DNA from synthesizing the WP complement at its 3' end. In the next high annealing temperature cycles, the target ssDNA is exponentially amplified because it is defined by both the WP and sequence-specific primer, while non-target ssDNA cannot be amplified as it lacks a binding site for at least one of the primers. Finally, the target DNA becomes the main PER-PCR product. This protocol has been validated by walking two selected genes.
Key features
• The current protocol builds upon the technique developed by Li et al. [1], which is universal to any species.
• The developed protocol relies on the partial overlap among a set of three WPs.
• Two rounds of nested PER-PCRs can generally result in a positive walking result.
Keywords: Genome walking (基因步移)Graphical overview
Background
In life science–related research, genome walking has achieved various applications, such as identifying transgenes, discovering new genetic resources, and cloning full-length genes, by mining unknown genomic regions flanking known DNA [2–4]. Therefore, genome walking has played an essential role in advancing life sciences–related disciplines like genetics, genomics, and microbiology [5–8].
At present, there are many genome walking methods available. The earliest genome walking method depended on constructing and then screening a genome library, which is laborious and time-consuming. Therefore, this method is seldom adopted now [9,10]. In contrast, PCR-based genome walking has received special attention due to its rapidity, low cost, and ease of operation [11–13]. To date, dozens of PCR-based genome walking methods have been established [14–17]. The existing PCR-based methods can be divided into two groups according to whether genome preprocessing is required preceding PCR: genome preprocessing-dependent PCR (such as panhandle PCR, inverse PCR, and restriction site extension PCR) and random PCR (such as wristwatch PCR, SiteFinding-PCR, and thermal asymmetric interlaced PCR). The latter has gradually become the mainstream walking method since it is free of the time-consuming genome preprocessing step. Although there are many random PCR walking methods available now, developing a practical analogous method is still popular [1,18].
Herein, a novel protocol based on primer extension refractory PCR (PER-PCR) is detailed for rapid and reliable genome walking. The PER-PCR relies on a set of three walking primers (WP): primary WP (PWP), secondary WP (SWP), and tertiary WP (TWP). The middle 15 nt of PWP overlaps the 3' part of SWP/TWP. The 5' parts of the three WPs are different from each other. The 15 nt overlap allows the WP to anneal to its predecessor site in the low annealing temperature (50 °C) cycle. However, the WP complement is unable to be produced on the DNA template due to the mismatch of the two WPs 5' regions. Clearly, only target DNA is amplified in the next 65 °C cycles. The disadvantage of PER-PCR is the complexity of designing a practical WP set. However, the three WP sets provided in this study are universal to any species. Users need only to design the required sequence-specific primer (SSP) set. This PER-PCR protocol can be used in many aspects, such as identifying new genes, unveiling regulatory DNAs, or clarifying structures of genetic elements [1].
Materials and reagents
Biological materials
1. Genomic DNA of rice was obtained from the lab of Dr. Xiaojue Peng at Nanchang University
2. Genomic DNA of Levilactobacillus brevis CD0817 [19–22] was extracted by our lab
Reagents
1. 10× LA PCR buffer (Mg2+ plus) (Takara, catalog number: RR042A)
2. 6× Loading buffer (Takara, catalog number: 9156)
3. LA Taq polymerase (hot-start version) (Takara, catalog number: RR042A)
4. dNTP mixture (Takara, catalog number: RR042A)
5. DL 5,000 DNA marker (Takara, catalog number: 3428Q)
6. 1× TE buffer (Sangon, catalog number: B548106)
7. Agarose (Sangon, catalog number: A620014)
8. 1 M NaOH (Yuanye, catalog number: B28412)
9. 0.5 M EDTA (Solarbio, catalog number: B540625)
10. Green fluorescent nucleic acid dye (10,000 ×) (Solarbio, catalog number: G8140)
11. Tris (Solarbio, catalog number: T8060)
12. Boric acid (Solarbio, catalog number: B8110)
13. DiaSpin DNA Gel Extraction kit (Sangon, catalog number: B110092)
14. Primers (Sangon)
PWP1: AAAGTAGTCATGTATCTGCGTCCTAGTC
PWP2: AAAGTAGTCATGTATCTGGCAGTCATAG
PWP3: AAAGTAGTCATGTATCTGTGCTGTCTGA
SWP: GTCGGTCTGGGTAGTCATGTATCTG
TWP: ACCCTGTGCCGTAGTCATGTATCTG
SSP1-gadR: TCCTTCGTTCTTGATTCCATACCCT
SSP2-gadR: CCATTTCCATAGGTTGCTCCAAGGTCA
SSP3-gadR: TAGGATACTGGCTAAAATGAATTAACTCGGAT
SSP1-hyg: GGAAGTGCTTGACATTGGGGAGT
SSP2-hyg: AAGACCTGCCTGAAACCGAACTGC
SSP3-hyg: CAAGGAATCGGTCAATACACTACATGGC
Solutions
1. 2.5× TBE buffer (see Recipes)
2. 0.5× TBE buffer (see Recipes)
3. 100 μM primer (see Recipes)
4. 10 μM primer (see Recipes)
5. 1.5% agarose gel (see Recipes)
Recipes
1. 2.5 × TBE buffer
| Reagent | Final concentration | Amount |
| 0.5 M EDTA solution | 5 mM | 10 mL |
| Tris | 225 mM | 27 g |
| Boric acid | 225 mM | 13.75 g |
| ddH2O | n/a | 980 mL |
| Total | n/a | 1,000 mL |
| Adjust pH to 8.3 with 1 M NaOH and then replenish the solution to 1,000 mL with ddH2O. | ||
2. 0.5× TBE buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 2.5× TBE buffer | 0.5× | 200 mL |
| ddH2O | n/a | 800 mL |
| Total | n/a | 1,000 mL |
3. 100 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Powdery primer | 100 μM | n/a |
| 1× TE buffer | 1× | Volume specified in the sheet of primer synthesis |
| Total | n/a | Volume specified in the sheet of primer synthesis |
Note: Dilute a portion of the 100 μM primer to prepare 10 μM primer and store the remaining portion at -80 °C.
4. 10 μM primer
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| 100 μM primer | 10 μM | 1 μL |
| 1× TE buffer | 1× | 9 μL |
| Total | n/a | 10 μL |
Note: Prepare extra volume of a 10 μM primer and divide it into multiple 1.5 mL microcentrifuge tubes; then, store the microcentrifuge tubes at -80 °C. Take one tube at a time and store it at -20 °C after use.
5. 1.5% agarose gel
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Agarose | 1.5% | 1.5 g |
| 0.5× TBE buffer | 0.5× | 100 mL |
| Green fluorescent nucleic acid dye (10,000×) | 1× | 10 μL |
| Total | n/a | 100 mL |
Laboratory supplies
1. 0.2 mL thin-wall PCR tubes (Kirgen, catalog number: KG2311)
2. 10 μL pipette tips (Sangon, catalog number: F600215)
3. 200 μL pipette tips (Sangon, catalog number: F600227)
4. 1,000 μL pipette tips (Sangon, catalog number: F630101)
5. 1.5 mL microcentrifuge tubes (Labselect, catalog number: MCT-001-150)
Equipment
1. PCR apparatus (Analtytikjena, model: Biometra TAdvanced)
2. Electrophoresis apparatus (Beijing Liuyi, model: DYY-6C)
3. Gel imaging system (Bio-Rad, model: ChemiDoc XRS+)
4. Microcentrifuge (Tiangen, model: TGear)
Software and datasets
1. Oligo 7 software (Molecular Biology Insights, Inc., USA)
2. DNASTAR Lasergene software (DNASTAR, Inc., USA)
Procedure
文章信息
稿件历史记录
提交日期: Sep 14, 2024
接收日期: Dec 12, 2024
在线发布日期: Dec 26, 2024
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Yu, Z., Wang, D., Lin, Z. and Li, H. (2025). Protocol to Mine Unknown Flanking DNA Using PER-PCR for Genome Walking. Bio-protocol 15(4): e5188. DOI: 10.21769/BioProtoc.5188.
分类
微生物学 > 微生物遗传学 > DNA > PCR
分子生物学 > DNA > PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link