- EN - English
- CN - 中文
Quantifying Bacterial Chemotaxis in Controlled and Stationary Chemical Gradients With a Microfluidic Device
利用微流控设备定量测量细菌在受控和静止化学梯度中的趋化性
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5184 浏览次数: 1693
评审: Hsih-Yin TanAnonymous reviewer(s)
Abstract
Chemotaxis refers to the ability of organisms to detect chemical gradients and bias their motion accordingly. Quantifying this bias is critical for many applications and requires a device that can generate and maintain a constant concentration field over a long period allowing for the observation of bacterial responses. In 2010, a method was introduced that combines microfluidics and hydrogel to facilitate the diffusion of chemical species and to set a linear gradient in a bacterial suspension in the absence of liquid flow. The device consists of three closely parallel channels, with the two outermost channels containing chemical species at varying concentrations, forming a uniform, stationary, and controlled gradient between them. Bacteria positioned in the central channel respond to this gradient by accumulating toward the high chemoattractant concentrations. Video-imaging of bacteria in fluorescent microscopy followed by trajectory analysis provide access to the key diffusive and chemotactic parameters of motility for the studied bacterial species. This technique offers a significant advantage over other microfluidic techniques as it enables observations in a stationary gradient. Here, we outline a modified and improved protocol that allows for the renewal of the bacterial population, modification of the chemical environment, and the performance of new measurements using the same chip. To demonstrate its efficacy, the protocol was used to measure the response of a strain of Escherichia coli to gradients of α-methyl-aspartate across the entire response range of the bacteria and for different gradients.
Key features
• The protocol is based on a previously proposed system [1] that we improved for higher throughput.
• Setup allowing a rapid quantification of motility and chemotaxis responses.
• Seventeen hours were required from the start of an E. coli culture to the measurements to obtain the chemotactic velocity under various chemical conditions.
Keywords: Microfluidics (微流控技术)Graphical overview
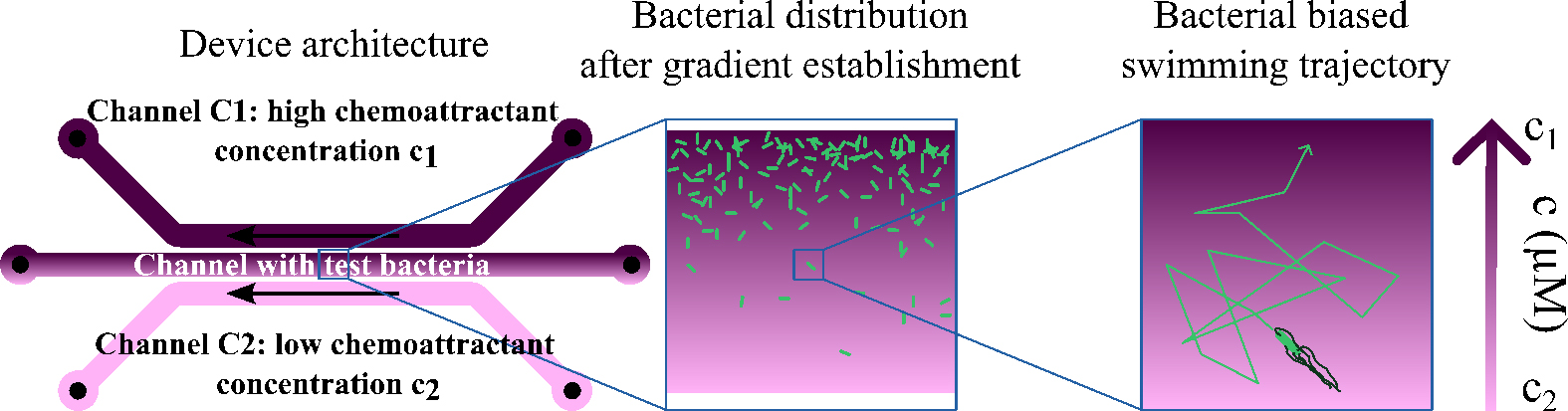
Schematic illustration of the three-channels chip architecture and its use with bacteria
Background
Bacterial chemotaxis is an important area of research that spans multiple fields. Examples include (i) microbiology, in which it helps to explain fundamental aspects of bacterial behavior, signaling pathways, and cellular processes; (ii) infectious diseases, where insights into how pathogenic bacteria use chemotaxis to locate and infect host tissues can inform the development of new treatments and preventive measures; (iii) environmental microbiology, in which chemotaxis is crucial for bacterial movement in natural environments, influencing nutrient cycling, bioremediation, and the functioning of microbial ecosystems; (iv) ecology and evolution, in which chemotaxis affects microbial interactions, competition, and cooperation within communities, providing insights into microbial ecology and the evolutionary dynamics of these behaviors; and (v) agriculture, where understanding how soil bacteria use chemotaxis to interact with plant roots can improve agricultural practices, enhance crop growth, and lead to the development of biofertilizers or biopesticides.
It is accepted that the chemotactic drift takes place at a velocity dependent on the detected concentration c and local gradient of the chemoattractant according to the following equation [2,3]:
(1)
where is the chemotactic susceptibility that depends on the local concentration
To this end, our microfluidic device uses diffusion through agar to generate a stable and controlled concentration and gradient and allows measuring precisely the vc for this environment. In practice, the gradient is established between parallel channels separated by a distance L, each containing a concentration cmin and cmax. Its value is and the average concentration of chemoattractant in which the bacteria swim is . Bacteria are introduced into the device, and their movement in response to the gradient is observed using microscopy. Bacteria swim toward the chemoattractant and gather on the channel wall closest to the source of the chemoattractant. Once the stationary regime is reached, the concentration of bacteria in this region decays exponentially, with a decay length:
(2)
This results from the competition between the advection of bacteria toward the source of chemoattractant at velocity vc and their active diffusion, which is characterized by a diffusion coefficient D. Here, the diffusion coefficient D is obtained by analyzing bacterial trajectories. This method enables the quantitative determination of chemotactic velocity and its dependence on the average concentration and its gradient. The chemotactic coefficient can be determined from the relation (see Eq. 1):
(3)
In its initial version, published by Ahmed et al. [1], the test required one microfluidic device per trial, making the method very time-consuming. Our protocol allows multiple trials to be run on the same chip, which may allow the method to be automated and enable continuous measurements. The method requires specialized equipment and expertise, which are presented here in detail. We illustrate the protocol using an Escherichia coli strain placed in α-methyl-aspartate gradients.
Materials and reagents
Biological materials
1. E. coli RP437 [7] (CGSC: #12122 transformed with the plasmid pZA3R-YFP carrying a chloramphenicol resistance marker and a yfp gene) stored in 25% glycerol in a -80 °C freezer for long-term conservation
Reagents
1. M9 salts (MP Biomedicals, catalog number: SKU 113037012-CF), dissolved in water at 22% (11 g in 50 mL)
2. Casamino acids (VWR, catalog number: ICNA113060012), dissolved in water at 5% (2.5 g in 50 mL)
3. D-(+)-Glucose (Sigma-Aldrich, catalog number: G7021-1KG), dissolved in water at 20% (10 g in 50 mL)
4. MgSO4·7H2O (Sigma-Aldrich, catalog number: 63138-250G), dissolved in water at 1 M (1.2 g in 50 mL)
5. KH2PO4 (Sigma-Aldrich, catalog number: P5655-500G), dissolved in water at 1 M (3.4 g in 25 mL water)
6. K2HPO4 (Sigma-Aldrich, catalog number: P3786-500G), dissolved in water at 1 M (4.3 g in 25 mL water)
7. CaCl2·2H2O (Sigma-Aldrich, catalog number: C3306-100G), dissolved in water at 1 M (36.8 mg in 50 mL)
8. Sodium lactate (Sigma-Aldrich, catalog number: L7022-10G), dissolved in water at 100 mM (1.1 g in 10 mL)
9. EDTA (Sigma-Aldrich, catalog number: EDS-100G), dissolved in water at 10 mM (292 mg in 10 mL)
10. L-methionine (Sigma-Aldrich, catalog number: M5308-25G), dissolved in water at 100 μM (149 mg in 1 L)
11. Chloramphenicol (Sigma-Aldrich, catalog number: C0378-5G)
12. Bacto agar (BD-Difco, catalog number: 214010)
13. PDMS components (Ellsworth, Dow Sylgard 184, GMID: 1673921)
a. Monomer
b. Curing agent
14. Ethanol 95% (Fisher Chemical, catalog number: E/0500DF/P21)
15. α-methyl-aspartate (MedChem, catalog number: HY-W142119)
Solutions
1. 100 mM KHPO4 buffer, pH 7 (see Recipes)
2. M9G medium (see Recipes)
3. Motility buffer (see Recipes)
4. PDMS (see Recipes)
5. 3% agar gel (see Recipes)
6. 70% ethanol (see Recipes)
7. Chloramphenicol stock solution (see Recipes)
Recipes
1. 100 mM KHPO4 buffer, pH 7
| Reagent | Final concentration | Amount |
|---|---|---|
| KH2PO4 (1 M) | 38.5 mM | 1.54 mL |
| K2HPO4 (1 M) | 61.5 mM | 2.46 mL |
| H2O | n/a | 36 mL |
| Total | n/a | 40 mL |
2. M9G medium
| Reagent | Final concentration | Amount |
|---|---|---|
| M9 salts (220 g/L) | 11.1 g/L | 25 mL |
| D-(+)-Glucose (200 g/L) | 4 g/L | 10 mL |
| Casamino acids (50 g/L) | 1 g/L | 10 mL |
| MgSO4·7H2O (100 mM) | 2 mM | 10 mL |
| CaCl2·2H2O (5 mM) | 100 μM | 10 mL |
| H2O | n/a | 435 mL |
| Total | n/a | 500 mL |
3. Motility buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| KHPO4 buffer (100 mM, pH 7.0) | 10 mM | 1 mL |
| Sodium lactate (1 M) | 10 mM | 100 μL |
| EDTA (100 mM, pH 10.0) | 100 μM | 10 μL |
| L-Methionine (1 mM) | 1 μM | 10 μL |
| H2O | n/a | 8.88 mL |
| Total | n/a | 10 mL |
4. PDMS
| Reagent | Final concentration | Amount |
|---|---|---|
| Monomer | 90% | 90 g |
| Curing agent | 9% | 9 g |
| Total | n/a | 99 g |
5. 3% agar gel
| Reagent | Final concentration | Amount |
|---|---|---|
| Bacto Agar | 30 g/L | 300 mg |
| H2O MilliQ | n/a | 10 mL |
| Total | n/a | 10 mL |
6. 70% ethanol
| Reagent | Final concentration | Amount |
|---|---|---|
| Ethanol (95%) | 70% | 350 mL |
| H2O | n/a | 125 mL |
| Total | n/a | 475 mL |
7. Chloramphenicol stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Chloramphenicol | 25 g/L | 250 mg |
| 70% ethanol | n/a | 10 mL |
| Total | n/a | 10 mL |
Store at -20 °C.
Laboratory supplies
1. Round-bottom double position 14 mL tubes (Falcon, catalog number: 352057)
2. 5 mL pipettes (Costar, Stripette, catalog number: 4487)
3. Micropipette tips (Gilson, D1000, catalog number: F167104)
4. Glass Petri dish D 120 mm, H 20 mm (Rogo Sampaic, catalog number: BRB005)
5. Scalpels (Swann Morton, catalog number: 0501)
6. Tubing (Darwin, Tygon, catalog number: LVF-KTU-13)
7. Safe-lock microtubes 2.0 mL (Eppendorf, catalog number: 0030120094)
8. Microvalve (Cluzeau CIL, catalog number: P-782)
9. 2 mm diameter sterile disposable biopsy punches (INTEGRA Miltex, catalog number: 33-31)
10. 2.5 mL syringes (Hamilton, Gastight 1002, catalog number: 81420)
11. Microscope glass slides 76 × 26 × 1 mm (Brand, catalog number: 474743)
12. Microscope glass slides 75.5 × 51.5 × 1.0 mm (Knittel, catalog number: VY11300051075.01)
13. Disposable plastic beaker 400 mL (Azlon, catalog number: BBBPB0400P) or paper cup
Equipment
1. Microscope (Leica, model: DMI 6000B, catalog number: 11888941)
a. Base (Leica, model: CTR6000, catalog number: 11888821)
b. Lens (Leica, model: HC PL FLUOTAR L 20×, catalog number: 11506243)
c. Fluorescence cube (Leica, model: Fluorescence Filter, Blue, I3, catalog number: 11513878): excitation filter: 450–490 μm; dichromatic mirror: 510 μm; suppression filter: 515 μm
d. Fluorescent light source (Leica, model: EL 6000, catalog number: 11504115)
e. Controller (Leica, model: IV/2013, catalog number: 11505180)
2. Digital CMOS camera (Hamamatsu, model: Orca-Flash4.0 V3, product number: C13440-20CU) mounted on the microscope
3. Syringe pump (Cetoni)
a. Base module (Cetoni Base 120, model: NEM-B100-01 F)
b. Three dosing units (Cetoni, Dosingmodule 14:1, model: NEM-B101-02 D)
Less sophisticated pumps can be used, including self-made pumps, see this link for example
4. Spectrophotometer (Eppendorf, model: D30, catalog number: 6133000001)
5. Pipette controller (Integra, model: PIPETBOY pro, catalog number: 156403/156401)
6. Biological safety cabinet (Telstar, model: Bio II Advance 3, catalog number: 523913)
7. Incubator shaker (Eppendorf, model: New Brunswick Innova 40R, catalog number: M1299-0086)
8. Autoclave (Advantage-Lab, model: AL02-01-100)
9. Oven [Labnet, catalog number: I5110(A)-230V]
10. Void Pump (KNF Lab, Laboport N816.3 KN.18, catalog number: 03533752)
11. Mini centrifuge (Sigma, model: 1-14, catalog number: 10014)
12. Stirrer hotplate (Fisher Scientific, catalog number: FB15001)
13. Semi-micro balance [Sartorius, model: CP(A)225D]
14. Precision balance (Sartorius, model: CP420S)
15. Vacuum desiccator (Bel-art, catalog number: F42022-0000)
16. Vortex mini mixer (Crystal LabPro, model: VM-03RUW)
17. -80 °C ultra-low temperature freezer (New Brunswick, New Brunswick Innova U101, model: U101-86)
18. Refrigerator: -20 °C freezer on top (Proline, DD133, catalog number: 7605749)
Software and datasets
1. NemeSys (v. 2016.06.14)
2. Leica Application SuiteX (v. 3.4.7)
3. Fiji (v. 1.52; https://imagej.net/software/fiji/downloads)
4. Python (v. 3.9; https://www.python.org/)
5. Inkscape (v. 1.3.2; https://inkscape.org/)
Procedure
文章信息
稿件历史记录
提交日期: Sep 11, 2024
接收日期: Dec 8, 2024
在线发布日期: Dec 31, 2024
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Gargasson, A., Douarche, C., Mergaert, P. and Auradou, H. (2025). Quantifying Bacterial Chemotaxis in Controlled and Stationary Chemical Gradients With a Microfluidic Device. Bio-protocol 15(4): e5184. DOI: 10.21769/BioProtoc.5184.
分类
生物信息学与计算生物学
生物科学 > 微生物学 > 微生物群落
微生物学 > 微生物生理学 > 趋化性
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link