- EN - English
- CN - 中文
Leveraging Circular Polymerization and Extension Cloning (CPEC) Method for Construction of CRISPR Screening Libraries
利用环状聚合酶扩展克隆(CPEC)方法构建CRISPR筛选文库
发布: 2025年02月20日第15卷第4期 DOI: 10.21769/BioProtoc.5183 浏览次数: 2772
评审: Kristin L. ShinglerEmmanuel Orta-ZavalzaAnonymous reviewer(s)

相关实验方案
在哺乳动物细胞中通过具有 3' 突出端的长 dsDNA 介导的 CRISPR 敲入 (LOCK) 实现高效的大 DNA 片段敲入
Wenjie Han [...] Jianqiang Bao
2023年10月20日 2770 阅读
Abstract
Recent advancements in high-throughput functional genomics have substantially enhanced our comprehension of the genetic and molecular dimensions of cancer, facilitating the identification of novel therapeutic targets. One of the key methodological innovations in this field is the CRISPR screening strategy, which has proven efficacy in elucidating essential gene functions and pathway alterations critical to cancer cell survival and fitness. The construction of custom CRISPR libraries permits the integration of tailored single-guide RNAs (gRNAs), offering greater flexibility as well as specificity in comparison to the commercially available libraries, and enables more refined secondary screening strategies to attenuate the selection of false positive potential gene candidates. Among various molecular cloning techniques, circular polymerase extension cloning (CPEC) has emerged as a highly efficient and cost-effective approach. CPEC utilizes polymerase overlap extension to assemble overlapping DNA fragments into circular plasmids, eliminating the need for restriction digestion and ligation and thus streamlining the creation of both single and multi-fragment constructs. In this protocol, we present the application of the CPEC method to construct the EpiTransNuc knockout gRNA library, specifically designed to target epigenetic regulators, transcription factors, and nuclear proteins. The custom library, assembled using the lentiGuide-Puro backbone, comprises 40,820 gRNAs, with 10 gRNAs per gene, along with 100 non-targeting control gRNAs. Importantly, the CPEC method can be tailored to meet the specific requirements of other custom gRNA libraries, offering flexibility for diverse research applications.
Key features
• Involves PCR-based linearization of the backbone with designed primer sets.
• Facilitates flexibility in gRNA composition and number in library construction.
• Skips conventional cloning techniques such as restriction digestion and ligation.
Keywords: Circular polymerase extension cloning (CPEC) (环状聚合酶扩展克隆(CPEC))Graphical overview
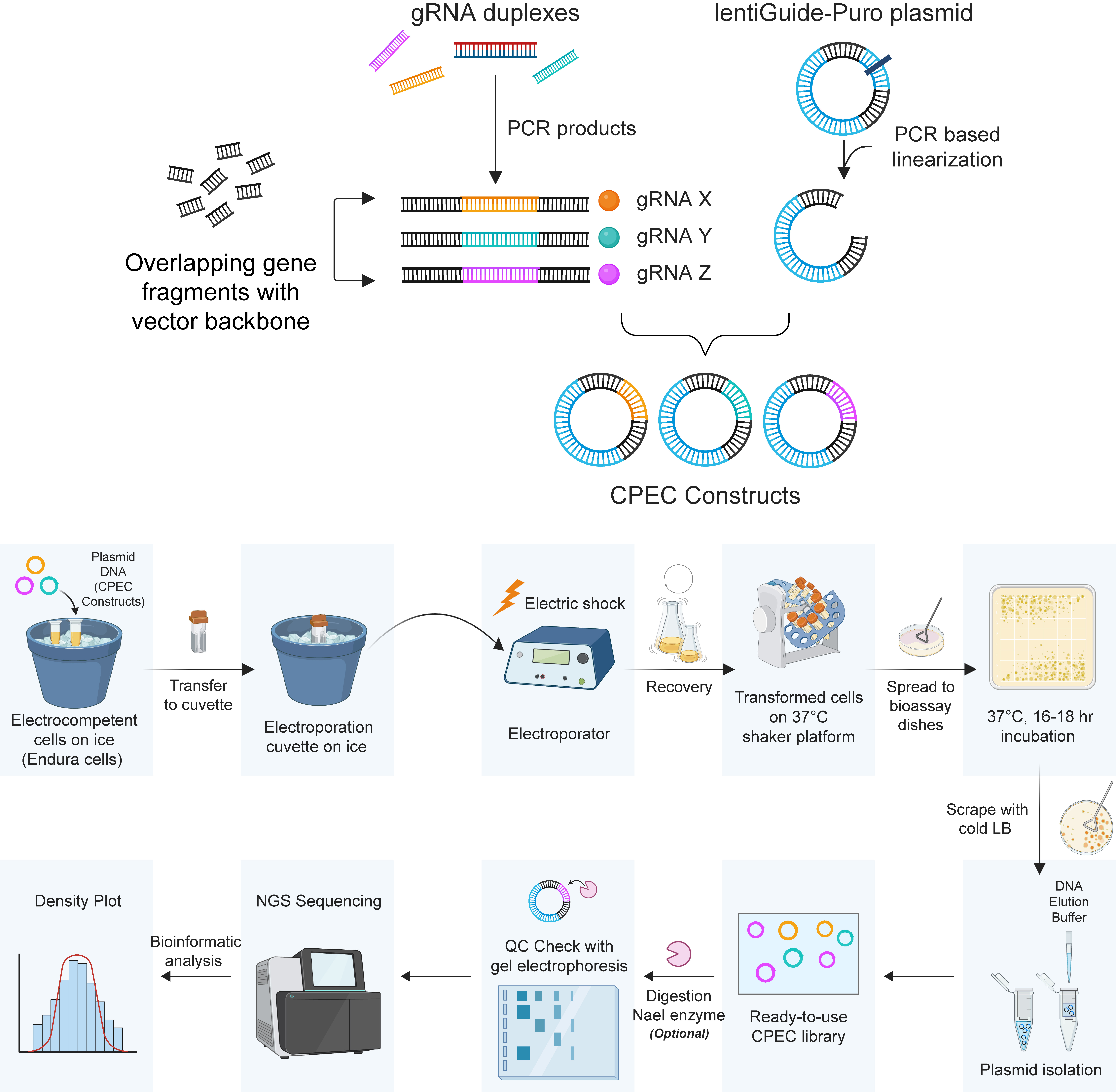
Schematic representation of the circular polymerase extension cloning (CPEC) procedure
Background
Our knowledge of the genetic and molecular aspects of cancer has advanced considerably owing to high-throughput functional genomics, which facilitates the identification of potential treatment targets or therapeutic vulnerabilities for a range of cancer types [1]. Genome-wide or focused CRISPR screening strategy has proven to be a versatile tool to unravel genes and even alterations in pathways having a major role in the survival and fitness of cancer cells, as well as therapy resistance [2,3]. Researchers can uncover details about gene functions, interactions, and regulatory networks by creating libraries containing different CRISPR constructs [4]. This understanding is crucial for deciphering complex biological systems and diseases.
Despite the availability of genome-wide or focused CRISPR libraries from commercial or repository sources, the creation of custom-built CRISPR libraries holds paramount importance for several reasons. First and foremost, de novo library construction enables the incorporation of a specific number of gRNAs, rather than being constrained by a predetermined or limited quantity. In contrast to genome-wide commercial libraries, which typically include a limited number of gRNAs per gene, custom-built libraries allow for a higher number of gRNAs per gene, thereby improving the reliability of the observed phenotypes. Moreover, to refine results and eliminate false negative candidates, secondary screens that focus on potential hits identified in primary screens are often conducted. To accomplish this, constructing a de novo library using gene sets that have already been pre-screened would become increasingly important. Second, the composition of the library can be adjusted with respect to the focus of the research. Although this study explores the CRISPR library within the context of cancer research, its application extends beyond this field. To exemplify, engineering gRNAs targeting transcription factors, metabolic genes, and immune regulation genes can be prioritized based on the specific research focus. Last but not least, a vector backbone with a Cas9 expression cassette or one that lacks Cas9 can be chosen.
In this context, researchers exploit cloning methodologies, which are classified into two primary categories: those that are sequence-dependent (Gateway cloning) and those that are sequence-independent [5], typically based on homologous recombination. Sequence-dependent strategies utilize restriction digestion-ligation methods or site-specific recombination and necessitate unique and specific sites within the insert, vector, or both. This dependence on unique sites makes these approaches less ideal for multi-fragment cloning. On the other hand, sequence-independent approaches comprise of ligation independent cloning (LIC) [6], sequence and ligation-independent cloning (SLIC) [7], Gibson assembly [8], and covalently-closed-circular synthesized (3Cs) [9]. All of these methods have pros and cons, including circular polymerization extension cloning (CPEC); however, CPEC could be exploited for the construction of CRISPR libraries whenever someone opts for a more cost-effective and streamlined approach.
Circular polymerase extension cloning (CPEC) represents a relatively cost-effective, high-efficiency cloning technique compared to the Gibson assembly and other protocols used in molecular cloning. It operates on the principle of polymerase overlap extension and is considered a robust alternative due to its omission of restriction digestion, ligation, and other procedural steps [10]. CPEC enables the transformation of overlapping DNA fragments into a double-stranded circular form via the polymerase extension mechanism, thereby facilitating the integration of the insert into the target plasmid. During the CPEC reaction, linear double-stranded inserts and vectors are first separated through increasing temperature (denaturation). Subsequently, the resulting single-stranded products anneal through their overlapping regions and use each other as templates to construct the circular plasmid. Through the CPEC method, not only a single gene but also multi-fragment assembly could be obtained. From this point of view, CPEC could have a pivotal role in the construction of CRISPR libraries (de novo). In this method, it is crucial to uniquely select the overlapping regions of the insert and vector, and the melting temperature (Tm) should be as high as possible to minimize the likelihood of vector self-ligation and concatenation.
Here, we present a robust, cost-effective, and straightforward CPEC-based protocol to engineer a custom-built gRNA library for CRISPR screening research. The knockout gRNA library, enriched for transcription factors and epigenetic regulators as well as nuclear factors, hence named EpiTransNuc, was constructed by exploiting the PCR linearized lentiGuide-Puro backbone and comprises 40820 gRNAs, comprising 10 gRNAs per gene and 100 non-targeting controls.
Materials and reagents
Biological materials
1. Endura electrocompetent E. coli bacteria (Lucigen, catalog number: 60242-1)
Reagents
1. lentiGuide-Puro backbone (Addgene, catalog number: 52963)
2. Designed primer sets for lentiGuide-Puro backbone linearization:
a. Forward primer: GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACC
b. Reverse primer: CGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGG
3. Q5 reaction buffer (New England Biolabs, catalog number: B9027S)
4. Q5 high GC enhancer (New England Biolabs, catalog number: B9028A)
5. 10 mM dNTPs (New England Biolabs, catalog number: N0447S)
6. Q5 high-fidelity DNA polymerase enzyme (New England Biolabs, catalog number: M0491S)
7. Ultra-pure DNase/RNase-free distilled water (Invitrogen, catalog number: 10-977-015)
8. NucleoSpin gel and PCR clean-up (Macherey-Nagel, catalog number: 740609.50)
9. EpiTransNuc library synthesized by CustomArray (Genscript) (the gRNA pool was improved upon the Addgene #51047 nuclear proteins gRNA sub-pool library through the inclusion of gRNAs from Sabatini, Brunello, GecKO, and Toronto v3)
10. Absolute ethanol (Isolab, catalog number: 9.200.262.500)
11. 10× FastDigest Esp3I (BsmBI, IIs class) enzyme (Thermo Fisher Scientific, catalog number: FD0454)
12. 10× FastDigest buffer (Thermo Fisher Scientific, catalog number: FD0454)
13. DTT (Neofroxx, catalog number: 1114GR005)
14. NEBNext high-fidelity PCR master mix (New England Biolabs, catalog number: M0541S)
15. Agarose Biomax (Prona, catalog number: 000320PR)
16. Gel loading dye, purple 6× (New England Biolabs, catalog number: B7024SVIAL)
17. 1 kb plus DNA ladder (New England Biolabs, catalog number: N3200L)
18. Isopropanol (Isolab, catalog number: 961.023.2500)
19. Electroporation cuvettes (Bio-Rad, catalog number:1652089)
20. Tryptone (AppliChem, catalog number: 1553.03)
21. Yeast extract (Nzytech, catalog number: MB16401)
22. NaCl (Merck, catalog number: M106404.1000)
23. LB broth with agar (Sigma-Aldrich, catalog number: L2897)
24. Ampicillin (Sigma-Aldrich, catalog number: A0166)
25. Tris base (Sigma-Aldrich, catalog number: T1503-1KG)
26. EDTA (Sigma-Aldrich, catalog number: E5134-500G)
27. Glacial acetic acid (Sigma-Aldrich, catalog number: 27225-2.5L-R)
28. SafeView Classic (ABM, catalog number: G108)
29. Endotoxin-free HiSpeed Plasmid DNA Maxi kit (Qiagen, catalog number: 12362)
Solutions
1. 10× Tris-acetate-EDTA buffer stock solution (10× TAE) (see Recipes)
2. Luria Bertani (LB) agar stock solution (see Recipes)
3. Luria Bertani (LB) liquid medium stock solution (see Recipes)
Recipes
Note 1: For the preparation of recipes, in-house ddH2O was used.
Note 2: A 10× TAE solution is used as stock and is stable at room temperature. Prepare LB agar stock and LB liquid medium stock solutions right before usage and keep them at 4 °C.
1. 10× Tris-acetate-EDTA buffer stock solution (10× TAE)
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tris base | 400 mM | 48.4 g |
| EDTA | 9.9 mM | 3.7 g |
| Glacial acetic acid | 1.14% (v/v) | 11.4 mL |
| ddH2O | - | 800 mL |
| Total | n/a | Complete with ddH2O to 1,000 mL |
2. LB agar stock solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| LB broth with agar | 3.5% (w/v) | 35 g |
| ddH2O | - | 1,000 mL |
| Total | n/a | 1,000 mL |
3. LB liquid medium stock solution
| Reagent | Final concentration | Quantity or Volume |
|---|---|---|
| Tryptone | 1% (w/v) | 10 g |
| Yeast extract | 0.5% (w/v) | 5 g |
| NaCl | 1% (w/v) | 10 g |
| ddH2O | 1,000 mL | |
| Total | n/a | 1,000 mL |
Laboratory supplies
1. Nunc square bioassay dishes (Thermo Fisher Scientific, catalog number: 240835)
2. 0.2 mL PCR tubes (Labselect, catalog number: PT-02-C)
3. 1.5 mL Eppendorf (Golden Gate, catalog number: KG2211)
4. Petri dish Ø90 × 17 mm (Isolab, catalog number: 081.02.191)
5. Drigalski glass spreader (Superior, catalog number: C180024)
Equipment
1. SimpliAmp thermal cycler (Applied Biosystems, model: A24811)
2. NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, model: ND-2000)
3. Microcentrifuge (Thermo Fisher Scientific, model: MicroCL 17R 230 V)
4. Balance (Sartorious, model: ENTRIS 822-1S)
5. Micropulser electroporator (Bio-Rad, catalog number: 1652100)
6. Gel electrophoresis system (Thermo Fisher Scientific, model: OWL B1A)
7. Gel Doc XR+ system with Image Lab software (Bio-Rad, model: 1708195)
8. 37 °C bacterial orbital shaker (Thermo Fisher Scientific, model: 4329)
9. 37 °C incubator (Heraeus, model: Heracell)
10. Heat block (Thermo Fisher Scientific, model: Digital Shaking Drybath)
11. Centrifuge (Eppendorf, model: 5810R)
Software and datasets
1. Python v2.7.17
2. Biopython v1.79
3. Count_spacers.py code (code is deposited at https://github.com/fengzhanglab/Screening_Protocols_manuscript)
4. BioRender (https://www.biorender.com/). The following figures were created using BioRender: Graphical overview, Senturk, S. (2025) https://BioRender.com/o79n165
Procedure
文章信息
稿件历史记录
提交日期: Sep 25, 2024
接收日期: Dec 8, 2024
在线发布日期: Dec 31, 2024
出版日期: Feb 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Dayanc, B., Eris, S. and Senturk, S. (2025). Leveraging Circular Polymerization and Extension Cloning (CPEC) Method for Construction of CRISPR Screening Libraries. Bio-protocol 15(4): e5183. DOI: 10.21769/BioProtoc.5183.
分类
分子生物学 > DNA > DNA 克隆
癌症生物学 > 通用技术 > 分子生物学技术
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link