- EN - English
- CN - 中文
Using the Sleeping Beauty Transposon System for Doxycycline-inducible Gene Expression in RAW264.7 Macrophage Cells to Study Phagocytosis
利用睡美人转座子系统在RAW264.7巨噬细胞中构建可四环素诱导的基因表达模型以研究吞噬作用
发布: 2025年02月05日第15卷第3期 DOI: 10.21769/BioProtoc.5178 浏览次数: 2002
评审: Paurvi ShindeChiara Ambrogio

相关实验方案

研究免疫调控血管功能的新实验方法:小鼠主动脉与T淋巴细胞或巨噬细胞的共培养
Taylor C. Kress [...] Eric J. Belin de Chantemèle
2025年09月05日 3542 阅读
Abstract
Macrophages are known for engulfing and digesting pathogens and dead cells through a specialized form of endocytosis called phagocytosis. Unfortunately, many macrophage cell lines are refractory to most reagents used for transient transfections. Alternative transient approaches, such as electroporation or transduction with lentiviral vectors, typically cause cell death (electroporation) or can be time-consuming to generate numerous lentivirus when using different genes of interest. Therefore, we use the Sleeping Beauty system to generate stably transfected cells. The system uses a “resurrected” transposase gene named Sleeping Beauty found in salmonid fish. Experimentally, the system introduces two plasmids: one carrying the Sleeping Beauty transposase and the other with an integration cassette carrying the gene of interest, a reverse-doxycycline controlled repressor gene, and an antibiotic resistance gene. The construct used in this protocol provides puromycin resistance. Stable integrations are selected by culturing the cells in the presence of puromycin, and further enrichment can be obtained using fluorescence-activated cell sorting (FACS). In this protocol, we use the Sleeping Beauty transposon system to generate RAW264.7 cells with doxycycline-inducible inositol polyphosphate 4-phosphatase B containing a C-terminal CaaX motif (INPP4B-CaaX). INPP4B-CaaX dephosphorylates the D-4 position of phosphatidylinositol 3,4-bisphosphate and inhibits phagocytosis. One benefit is that generating stable cell lines is substantially faster than selecting for random integrations. Without FACS, the method typically gives ~50% of the cells that are transfected; with sorting, this approaches 100%. This makes phagocytosis experiments easier since more cells can be analyzed per experiment, allowing for population-based measurements where a ~10% transient transfection rate is insufficient. Finally, using the doxycycline-promoter allows for low near endogenous expression of proteins or robust overexpression.
Key features
• This protocol builds on the protocols and reagents developed by Kowarz et al. [1] and extends it to using RAW macrophages.
• Allows for the rapid generation of stably induced cell lines.
• This protocol also determines the phagocytic index and efficiency.
Keywords: Phagocytosis (吞噬作用)Graphical overview
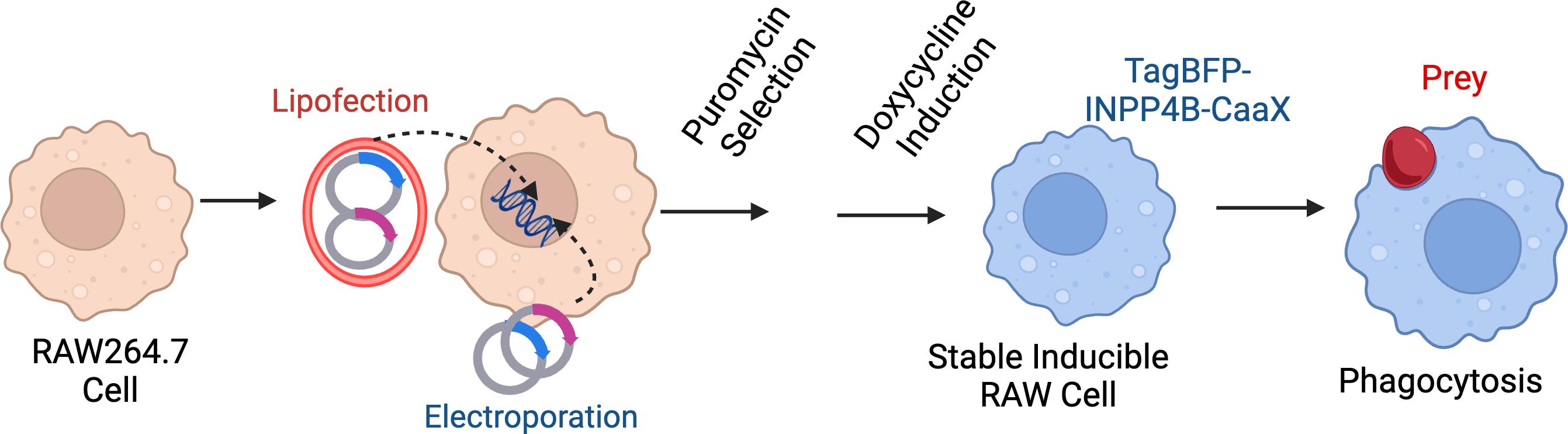
Protocol overview. Through electroporation or lipofection, RAW 264.7 cells are transfected with two plasmids: one carrying the Sleeping Beauty transposase and the other with an integration cassette carrying the gene of interest, reverse doxycycline-controlled repressor gene, and a puromycin-resistance gene. Transfected cells are then selected with 2.5 μg/μL of puromycin for at least 5 days to select for a polyclonal mix of stably transfected cells. For experiments, cells are treated with doxycycline to induce the TagBFP-INPP4B-CAAX or the inactive control TagBFP-INPP4BC842A-CAAX. Cells are exposed to opsonized particles to induce phagocytosis.
Background
The introduction of plasmid DNA into mammalian cells has been an essential method to support cellular and molecular biology investigations. Typically, the introduction of plasmid DNA can be mediated by a few approaches, including electroporation, chemical transformation, polyfection using polymer-based transfection reagents, microinjection, and viral transduction. These methods vary in efficiency depending on cell type and scale. Another factor to consider is whether the researcher wants transient over-expression of a gene of interest or generation of stably transfected cell lines. The generation of a stable cell line is typically more time-consuming. Still, it can be advantageous if cells are refractory to lipofection or if the same cell lines will be used for several experiments. In our lab, we use the Sleeping Beauty transposon system to help facilitate the integration of DNA into the genome to support the generation of stable cell lines [2–4]. This transposon system has been widely used as a genetic engineering tool for the last decade. Indeed, we have used this system to generate RAW264.7 cells expressing the reverse tetracycline repressor (rTetR) for controllable induction of our protein of interest, TagBFP-INPP4B-CaaX [5]. As these cells are generally challenging to transfect transiently, this approach allows for the rapid creation of cell lines.
The protocol described here is an adaptation of Kowarz et al. [1]. Briefly, this system utilizes a plasmid (SB100x) encoding the Sleeping Beauty transposase and a second plasmid containing an integration cassette with a gene of interest, a selectable marker, flanked by inverted terminal repeats. Kowarz et al. [1] generated a variety of plasmids, available from Addgene, to allow researchers to create cells with constitutive or doxycycline-inducible genes, an array of drug-resistant markers (e.g., puromycin or hygromycin) and the option also to deliver the reverse tetracycline repressor.
Phagocytosis is an actin-driven process used when cells ingest particles greater than 0.5 mm, such as pathogens and apoptotic bodies [6–8]. Phagocytosis entails the extension of actin-rich pseudopods to surround the target and pull the particle into the cell, forming a nascent phagosome [9]. The extensive actin remodeling, polymerization, bundling, and depolymerization must be exquisitely controlled and temporally coordinated. Major regulators in this process are the metabolism and interconversion of phosphoinositides lipids [10]. For instance, the generation of phosphatidylinositol (3,4)-bisphosphate (PtdIns3,4P2) at the engagement site is crucial for extending pseudopods to envelop prey [5]. Mammalian cells contain at least two pathways and multiple enzymes capable of synthesizing PtdIns3,4P2; thus, the knockdown of individual enzymes or specific inhibitors often generates unsatisfying results [11]. Instead, we used a synthetic plasmid-borne enzyme inositol polyphosphate 4-phosphatase type IIB (INPP4B) produced as a chimera with TagBFP for visualization, as well as a C-terminal CaaX motif for prenylation and membrane targeting [12]. This chimeric protein catalyzes the dephosphorylation of the D-4 position of PtdIns3,4P2 in the plasma membrane regardless of the biosynthetic route [12].
We generate doxycycline-inducible RAW264.7 cells expressing BFP-INPP4B-CaaX and a second cell line expressing the catalytically inactive BFP-INPP4BC842A-CaaX that serves as a control. Following overnight treatment with doxycycline to induce the INPP4B-containing constructs, we conduct phagocytosis assays to determine the role of PtdIns3,4P2 [5]. Typically, our phagocytosis assays use IgG-opsonized sheep red blood cells or IgG-coated polystyrene beads as prey to assess Fcg receptor-mediated phagocytosis [13].
Many approaches are available to assess phagocytosis; most rely on fluorescent labels on the prey to quantify internalization and phagosome maturation. Flow cytometry analysis determines the overall ingestion of particles by measuring total fluorescence intensity/cell as a parameter for particle uptake [14]. Other methods use pH-sensitive fluorescent particles that become brighter in acidic conditions, such as in the phagolysosome, that measure particle uptake and phagosome maturation [15]. However, both these approaches are limited in defining particle engagement vs. complete engulfment. Instead, we prefer a high-resolution confocal microscope and an antibody staining approach to distinguish bound vs. fully internalized particles (Figure 1).
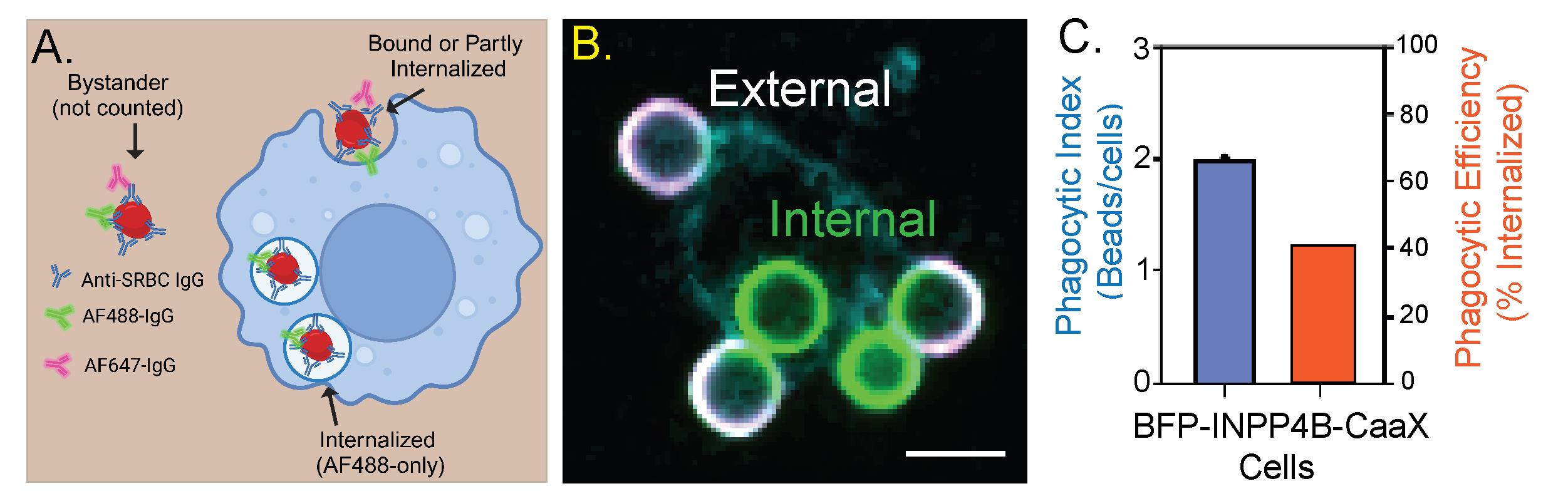
Figure 1. INPP4B-CaaX inhibits the internalization of IgG opsonized particles. A. Representation of the assay. All sheep red blood cells (SRBCs) are labeled with the AF488-conjugated antibody, whereas only extracellular SRBCs are stained with the AF647 antibody at the end of the incubation period. B. Representative image. Fluorescence markers are pseudocolored: AF488 (green), AF647 (magenta), and BFP (cyan). Scale bar = 5 μm. C. A double y-axis graph depicting the results of panel B. The phagocytic index represents the number of beads internalized by the cells over a fixed period. Phagocytic efficiency represents the percentage of engaged particles that are internalized.
Materials and reagents
Biological materials
1. RAW264.7 cell line (American Type Culture Collection, catalog number: TIB-71)
2. Sheep red blood cells, 10% suspension (MP Biomedicals, catalog number: 55876), stored at 2–8 °C
3. pCMV(CAT)-T7-SB100X plasmid (Addgene, plasmid #34879)
4. pSBtet-Pur plasmid (Addgene, plasmid #60507)
5. Primary antibodies (store at -20 °C):
a. Anti-sheep red blood cell rabbit IgG antibody (85 mg/mL) (Rockland, catalog number: 113-4139)
b. Total human serum IgG antibody (50 mg/mL) (Millipore Sigma, catalog number: I4381)
6. Secondary antibodies (store at -20 °C):
a. Goat anti-rabbit IgG (H+L) antibody-AlexaFluor647 (Jackson ImmunoResearch, catalog number: 111-605-144)
b. Goat anti-rabbit IgG (H+L) antibody-AlexaFluor488 (Jackson ImmunoResearch, catalog number: 111-545-144)
c. Donkey anti-human IgG (H+L) antibody-AlexaFluor647 (Jackson ImmunoResearch, catalog number: 709-605-149)
d. Donkey anti-human IgG (H+L) antibody-AlexaFluor488 (Jackson ImmunoResearch, catalog number: 709-545-149)
Reagents
1. Paraformaldehyde 16% wt/vol (PFA) (Electron Microscopy Sciences, catalog number: 15700)
2. Doxycycline hyclate (Sigma-Aldrich, catalog number: D9891)
3. Puromycin (Sigma-Aldrich, catalog number: P8833
4. Roswell Park Memorial Institute Medium (RPMI) 1640 with L-glutamine and sodium pyruvate (Wisent Bioproducts, catalog number: 350-015-CL)
5. Phosphate buffered saline (PBS) without calcium and magnesium (Wisent bioproducts, catalog number: 311-013-CL)
6. Phosphate buffered saline with calcium and magnesium (PBS+/+) (Wisent bioproducts, catalog number: 311-011-CL)
7. Fetal bovine serum (FBS) (Wisent Bioproducts, catalog number: 080-150)
8. 4.2 mm Polystyrene beads with 2% divinylbenzene (Bangs Laboratories, Inc., catalog number: PS06005), stored at 2–8 °C
9. Fugene HD (Promega, catalog number: E2311)
10. Electrolytic buffer E2 (Invitrogen, catalog number: MPK10096E)
11. Resuspension buffer R (Invitrogen, catalog number: MPK10096R)
Solutions
1. 4% PFA (see Recipes)
2. Selection medium (see Recipes)
3. Induction medium (see Recipes)
Recipes
1. 4% PFA
| Reagent | Final concentration | Amount |
|---|---|---|
| 16% PFA | 4% | 1 mL |
| PBS+/+ | n/a | 3 mL |
| Total | n/a | 4 mL |
2. Selection medium (2.5 μg/mL of puromycin in RPMI)
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | n/a | ~10 mL |
| Puromycin (10 mg/mL) | 2.5 μg/mL | 2.5 μL |
| Total | n/a | 10 mL |
3. Induction medium (1 μg/mL of doxycycline in RPMI)
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | n/a | ~10 mL |
| Doxycycline hyclate (10 mg/mL) | 1 μg/mL | 2.5 μL |
| Total | n/a | 10 mL |
Laboratory supplies
1. T-25 (Sarstedt, catalog number: 83.3910)
2. T-75 (Sarstedt, catalog number: 83.3911)
3. Cell scraper (Wuxi NEST Biotechnology, catalog number: 710001)
4. 6-well plates (Sarstedt, catalog number: 83.3920)
5. 12-well plates (Sarstedt, catalog number: 83.3921)
6. 18 mm circular cover glass, #1½ (Electron Microscopy Sciences, catalog number: 72222-01
7. Kimwipes (Kimberly-Clark ProfessionalTM 34155, catalog number: 06-666A)
8. 1.5 mL Eppendorf tubes (Froggabio, catalog number: LMCT1.7B)
9. 15 mL tubes (Fisher Scientific, catalog number: 14-959-53A)
Equipment
1. Incubator (Eppendorf, model: CellXpert C170, catalog number: 6734)
2. Tabletop centrifuge (Thermo Scientific, model: Pico 21, catalog number: 75002553)
3. Plate centrifuge (Thermo Scientific, model: Sorvall Legend RT, catalog number: 75004377)
4. Tube revolver (Thermo Scientific, catalog number: 88881001)
5. Vortex (Scientific Industries, catalog number: SI-0236)
6. Neon Invitrogen transfection system (Thermo Fisher Scientific, Invitrogen, catalog number: MPK5000)
7. Hemocytometer (NanoEntek, catalog number: EVE-MC)
8. 3i MarianasTM spinning-disc confocal microscopy, based on the Zeiss Axio Observer 7 Advanced Microscope with Definite Focus 3 [Intelligent Imaging Innovation (3i), custom-built]
Software and datasets
The microscope and image acquisition were controlled with SlideBook 2024 (3i). TIFF images were exported and analyzed in ImageJ2 Version 2.14.0/1.54f [16]. Representative images were chosen based on a good signal-to-noise ratio. Merging and cropping fluorescent channels were performed in ImageJ2. To aid visualization, linear adjustments were made to brightness and contrast across the entire image. To help with data accessibility and enhance the presentation of micrographs, we switched the default red lookup table to magenta. Graphical overview was generated using BioRender.com. Figure 1 was assembled using Adobe Illustrator 2024 and GraphPad Prism V9.
Procedure
文章信息
稿件历史记录
提交日期: Sep 30, 2024
接收日期: Dec 2, 2024
在线发布日期: Dec 26, 2024
出版日期: Feb 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kamali, P. and Fairn, G. D. (2025). Using the Sleeping Beauty Transposon System for Doxycycline-inducible Gene Expression in RAW264.7 Macrophage Cells to Study Phagocytosis. Bio-protocol 15(3): e5178. DOI: 10.21769/BioProtoc.5178.
- Montaño-Rendón, F., Walpole, G. F., Krause, M., Hammond, G. R., Grinstein, S. and Fairn, G. D. (2022). PtdIns(3,4)P2, Lamellipodin, and VASP coordinate actin dynamics during phagocytosis in macrophages. J Cell Biol. 221(11): e202207042.
分类
免疫学 > 免疫细胞功能 > 巨噬细胞
细胞生物学 > 基于细胞的分析方法 > 基因表达
免疫学 > 免疫机理
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link