- EN - English
- CN - 中文
Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology
靶向纳米孔测序技术在结核分枝杆菌及其耐药性检测中的应用
发布: 2025年02月05日第15卷第3期 DOI: 10.21769/BioProtoc.5170 浏览次数: 1978
评审: Alka MehraSoumya MoonjelySuresh Panthee
Abstract
Tuberculosis (TB) remains the leading cause of human mortality in infectious diseases. Drug-resistant TB, particularly multidrug-resistant TB and extensively drug-resistant TB, poses a pressing clinical and public health challenge. The main causative agents of TB are known as Mycobacterium tuberculosis (MTB), which exhibits a highly complex drug resistance profile. Traditional culture-based phenotypic drug susceptibility testing is time-consuming, and PCR-based assays are restricted to detecting known mutational hotspots. In this study, we present a protocol leveraging high-throughput nanopore sequencing technology in conjunction with multiplex PCR, termed targeted nanopore sequencing, for the identification of MTB and analysis of its drug resistance. Our method for MTB drug resistance assessment offers the benefits of being culture-free, efficient, high-throughput, and highly accurate, which could significantly aid in clinical patient management and the control of TB infections.
Key features
• Targeted nanopore sequencing detects 18 genes simultaneously linked to antibiotic resistance in MTB.
• The method provides broad drug resistance profiles for 14 first- and second-line anti-TB drugs without bacterial culture.
• The expedited turnaround time of the process is approximately 7.5 h with a detection limit of 102 bacteria/mL.
Keywords: Mycobacterium tuberculosis (结核分枝杆菌)Graphical overview
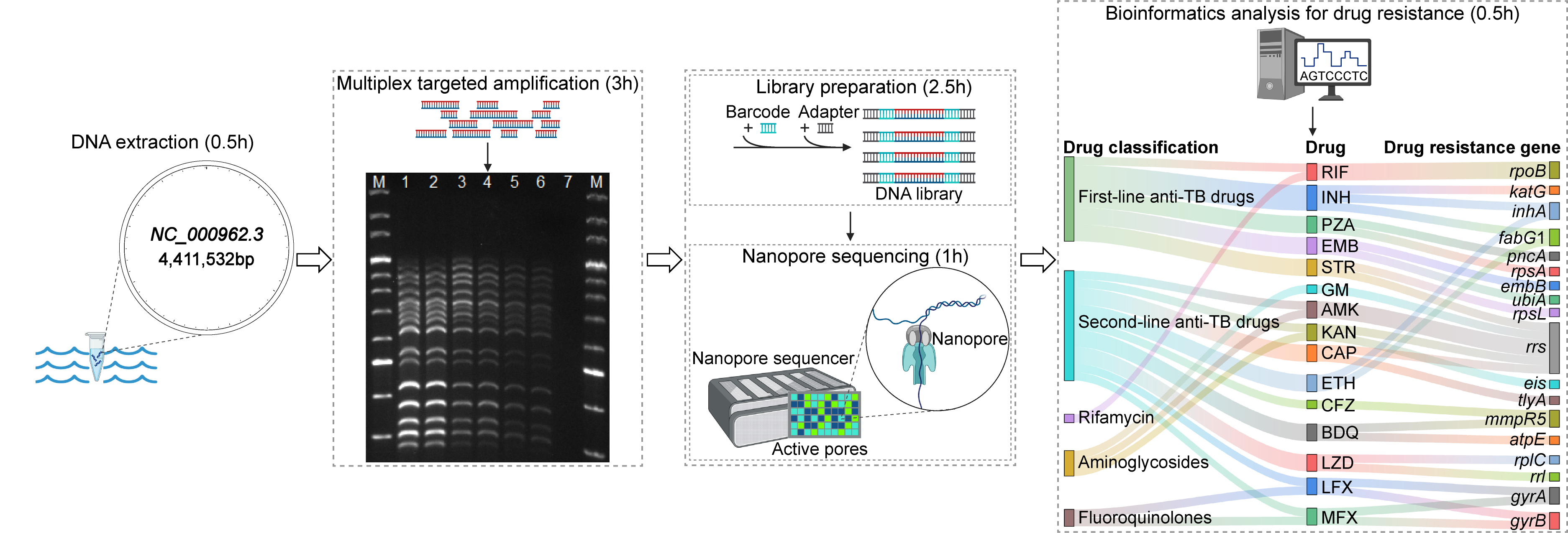
Targeted nanopore sequencing for the drug resistance assay of Mycobacterium tuberculosis. Initially, DNA is extracted, and the targeted regions within genes associated with drug resistance are amplified via multiplex PCR. Subsequently, the amplicons undergo barcoding and adapter ligation. The library preparation is then sequenced on a nanopore sequencer, which yields long-read sequence data. Ultimately, drug resistance is identified through bioinformatics analysis. The entire targeted nanopore sequencing process has an expedited turnaround time of approximately 7.5 h.
Background
Tuberculosis (TB) affirms its position as the preeminent cause of human death from infectious diseases, with an estimated 1.7 million deaths occurring globally each year, as reported by Daley [1]. The latest figures from the World Health Organization’s Global Tuberculosis Report 2023 reveal that 7.5 million new cases were diagnosed worldwide in 2022, surpassing pre-COVID levels. TB pathogen refers to a group of bacteria within the Mycobacterium tuberculosis complex (MTBC), including Mycobacterium tuberculosis (MTB), noted for its intricate drug resistance spectrum [2]. MTB, being the principal causative agent of TB, gains antibiotic resistance through gene mutations, significantly influencing clinical treatment approaches [3–5].
Surveillance of highly virulent and multidrug-resistant (MDR) MTB strains is imperative for securing precise diagnoses and potent treatment regimens [6]. Conventional culture-based phenotypic drug susceptibility testing (pDST) is the standard method for assessing MTB, although being time-consuming, typically requiring a span of days to weeks [3]. The prevalent molecular detection methods, including the real-time PCR-based Xpert MTB/RIF assay endorsed by the WHO, while being efficient, are limited to detecting known mutational hotspots and thus provide incomplete data on drug resistance [7]. Consequently, infections with MDR and extensively drug-resistant (XDR) MTB strains highlight an urgent need for advanced diagnostic tools to guide TB treatment options [8].
High-throughput sequencing technology has become a staple in clinical diagnostics. Nanopore sequencing, a cutting-edge method exemplified by the equipment from Oxford Nanopore Technologies (ONT), offers the benefits of long-read lengths, real-time data acquisition, and rapid sequencing [9]. While its accuracy has been a subject of improvement, significant strides in scientific and technological advancements have enhanced its performance in recent years [10,11]. Nanopore sequencing facilitates direct analysis of DNA or RNA sequences without the need for culturing MTB, enabling swift identification of MTB and detection of TB drug resistance. This approach might not only reduce the overall cost of MTB detection but also expedite the diagnostic process [12]. Therefore, the technology holds considerable promise for clinical applications and possesses a promising market outlook, making it an attractive option for clinical settings [13].
This study introduces a method that integrates multiplex PCR for targeted enrichment with nanopore sequencing, termed targeted nanopore sequencing. Furthermore, the bioinformatic tool TBProfiler is employed for drug resistance analysis, with its database encompassing 1,195 candidate mutations, which were obtained globally from more than 17,000 clinical isolates and assessed with whole-genome sequencing (WGS) and pDST [14].
Materials and reagents
Reagents
1. 10× TBE buffer (Solarbio, catalog number: T1051)
2. 1 M Tris-HCl (pH 8.0) (Solarbio, catalog number: T1150)
3. 200 bp DNA ladder (Dye Plus) (Takara, catalog number: 3423A)
4. Agarose (Mei5 Biotechnology, Co., Ltd, catalog number: MF103)
5. Agencourt AMPure XP beads (Beckman Coulter, catalog number: A63881)
6. Chelex 100 sodium (Solarbio, catalog number: C8230)
7. DNase/RNase-free water (Solarbio, catalog number: R1600)
8. dNTP mixture (Takara, catalog number: 4030)
9. EDTA (Sigma, catalog number: 798681)
10. Ligation Sequencing kit (Q20+) (Oxford Nanopore Technologies, catalog number: SQK-LSK114)
11. M5 6× DNA electrophoresis loading buffer (Mei5 Biotechnology, Co., Ltd, catalog number: MF144-01)
12. M5 Hipure Next III Gelred (Mei5 Biotechnology, Co., Ltd, catalog number: MF380-01)
13. NaOH (Sigma, catalog number: 221465)
14. Native Barcoding kit 24 (Q20+) (Oxford Nanopore Technologies, catalog number: SQK-NBD114.24)
15. NEB Blunt/TA ligase master mix (New England Biolabs, catalog number: M0367L)
16. NEB Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs, catalog number: M0493L)
17. NEBNext Ultra II end repair/dA-tailing module (New England Biolabs, catalog number: E7546S)
18. NEBNext quick ligation module (New England Biolabs, catalog number: E6056S)
19. Nonidet P40 substitute (NP-40) (Solarbio, catalog number: N8030)
20. PBS (Sigma, catalog number: P3813)
21. Qubit dsDNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q32854)
22. Triton X-100 (Solarbio, catalog number: T8200)
Laboratory supplies
1. 0.2 mL PCR tubes (ideally in strips of 8) (Thermo Fisher Scientific, catalog number: AB0490)
2. 1.5 mL tubes (Thermo Fisher Scientific, catalog number: S348903)
3. Pipette tips (Thermo Fisher Scientific, catalog numbers: 02-707-426, 02-707-403, 02-707-438)
4. PromethION flow cell R10.4.1 (Oxford Nanopore Technologies, catalog number: FLO-PRO114M)
5. Qubit assay tubes (Thermo Fisher Scientific, catalog number: Q32856)
Equipment
1. Biological safety cabinet (NuAire Lab Equipment, catalog number: NU-425-300)
2. Electrophoresis unit of power supply (Thermo Fisher Scientific, catalog number: S65533Q)
3. Magnetic stand for 1.5 mL tubes (Beckman Coulter, catalog number: A29182)
4. Microcentrifuge (Eppendorf, catalog number: EP5401000137)
5. Milli-Q ultrapure water system (Millipore Synergy, catalog number: F1CA45528 A)
6. NanoDrop spectrophotometer (Thermo Fisher Scientific, catalog number: ND-1000)
7. Qubit 4 fluorometer (Thermo Fisher Scientific, catalog number: Q33238)
8. Sequencing device (Oxford Nanopore Technologies, model: PromethION)
9. S1000 thermal cycler (Bio-Rad, catalog number: 1852196)
10. Vortex mixer (Thermo Fisher Scientific, catalog number: S96461A)
Software and datasets
1. Albacore (https://github.com/Albacore/albacore)
2. Bowtie2 (https://github.com/BenLangmead/bowtie2)
3. Delly (https://github.com/dellytools/delly)
4. MinKNOW (https://github.com/nanoporetech/minknow_api)
5. NanoFilt (https://github.com/wdecoster/nanofilt)
6. SAMtools (https://github.com/samtools/samtools)
7. TBProfiler (https://github.com/jodyphelan/TBProfiler)
8. Trimmomatic (https://github.com/usadellab/Trimmomatic)
Procedure
文章信息
稿件历史记录
提交日期: Jul 4, 2024
接收日期: Nov 28, 2024
在线发布日期: Dec 15, 2024
出版日期: Feb 5, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Tang, C., Xu, F., Zheng, X. and Xiang, G. (2025). Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology. Bio-protocol 15(3): e5170. DOI: 10.21769/BioProtoc.5170.
分类
微生物学 > 微生物遗传学 > DNA > DNA 测序
分子生物学 > DNA > DNA 测序
微生物学 > 抗微生物试验 > 抗细菌试验
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link