- EN - English
- CN - 中文
Using Protein Painting Mass Spectrometry to Define Ligand Receptor Interaction Sites for Acetylcholine Binding Protein
利用蛋白涂层质谱技术解析乙酰胆碱结合蛋白的配体受体相互作用位点
发布: 2025年01月20日第15卷第2期 DOI: 10.21769/BioProtoc.5163 浏览次数: 1915
评审: Sébastien GillotinSarajo MohantaRohit Jain

相关实验方案
基于蛋白筛选策略从合成文库中分离抗原特异性纳米抗体:结合 MACS 的酵母展示筛选与 FLI-TRAP 方法
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
2026年01月20日 408 阅读
Abstract
Nicotinic acetylcholine receptors (nAChRs) are a family of ligand-gated ion channels expressed in nervous and non-nervous system tissue important for memory, movement, and sensory processes. The pharmacological targeting of nAChRs, using small molecules or peptides, is a promising approach for the development of compounds for the treatment of various human diseases including inflammatory and neurogenerative disorders such as Alzheimer’s disease. Using the Aplysia californica acetylcholine binding protein (Ac-AChBP) as an established structural surrogate for human homopentameric α7 nAChRs, we describe an innovative protein painting mass spectrometry (MS) method that can be used to identify interaction sites for various ligands at the extracellular nAChR site. We describe how the use of small molecule dyes can be optimized to uncover contact sites for ligand–protein interactions based on MS detection. Protein painting MS has been recently shown to be an effective tool for the identification of residues within Ac-AChBP involved in the binding of know ligands such as α-bungarotoxin. This strategy can be used with computational structural modeling to identify binding regions involved in drug targeting at the nAChR.
Key features
• Identify binding ligands of nicotinic receptors based on similarity with the acetylcholine binding protein.
• Can be adapted to test various ligands and binding conditions.
• Mass spectrometry identification of specific amino acid residues that contribute to protein binding.
• Can be effectively coupled to structural modeling analysis.
Keywords: Ligand binding (配体结合)Graphical overview
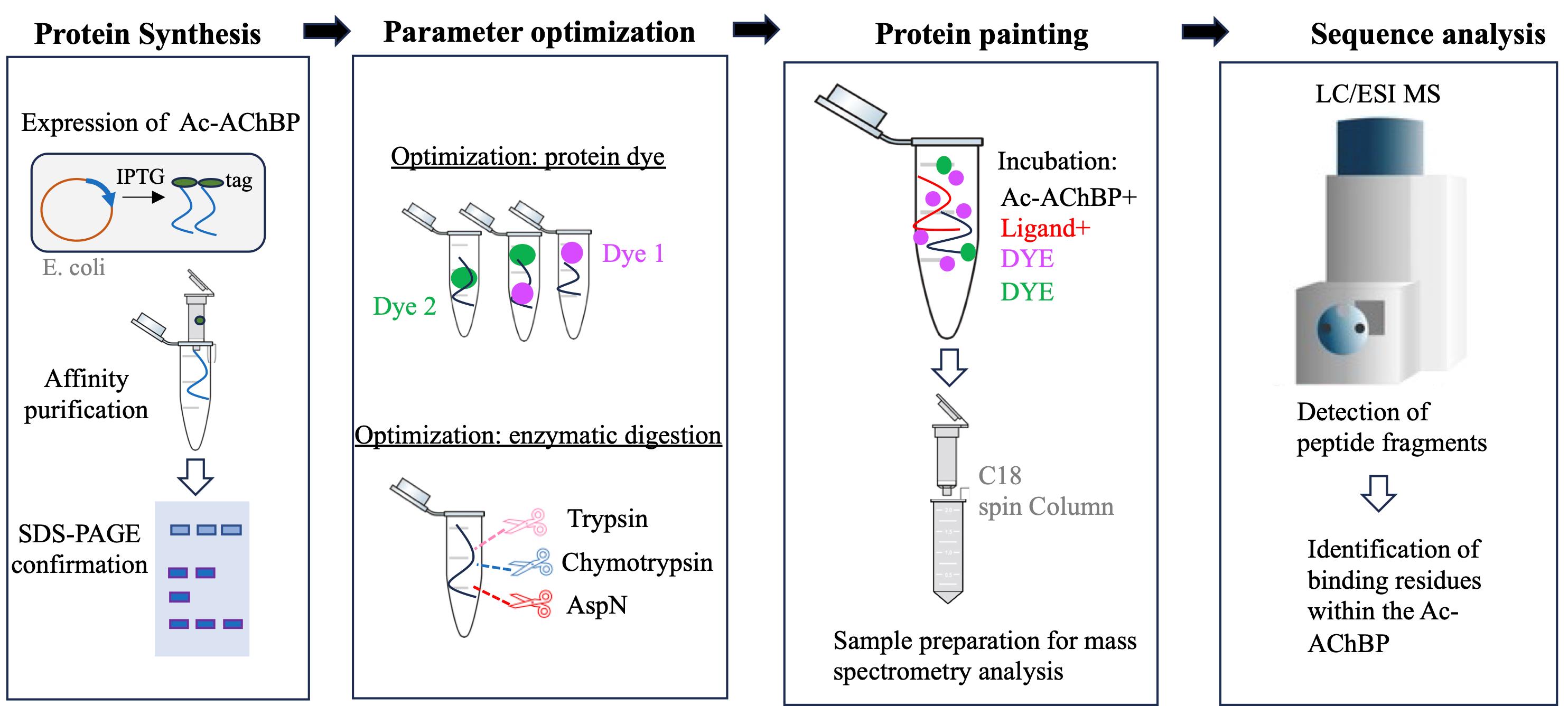
A summary of the workflow and main steps of the protein painting experimental design. Primary steps in the development and execution of the experiment. From left to right: The Ac-AChBP is produced in E. coli and then isolated using affinity chromatography; various dyes and enzymes are optimized prior to the protein paint experiment. Mass spectrometry analysis is used to define peptide fragments that are differentially produced in the painted experiment.
Background
Mammalian nicotinic acetylcholine receptors (nAChRs) are a family of ligand ion channels, which are widely expressed in the central and peripheral nervous systems. They play key roles in modulating neurotransmitter release, synaptic plasticity, and cognitive functions; they also regulate autonomic functions by mediating fast synaptic transmission in autonomic ganglia, impacting heart rate and blood pressure [1]. The pharmacological targeting of human nAChRs is a leading strategy for therapeutic drug development in the treatment of various nervous system disorders, including Alzheimer’s and neuropathic disease [2]. Homopentameric α7 nAChRs bind important proteins including pathogenic beta amyloids and various neurotoxins [3,4]. The structural similarity, solubility, and ligand-binding properties of the homopentameric invertebrate acetylcholine binding protein (AChBP) make it a suitable model for studying ligand binding at the extracellular domain of homopentameric nAChRs [5]. In this method paper, we describe the development of a protein painting mass spectrometry (MS) approach that can serve in the detection of protein binding domains within the AChBP.
Protein painting MS technology is a new biochemical strategy in the study of ligand binding to nAChRs, recently demonstrated by our study using AChBP derived from Aplysia californica (Ac-AChBP) [6,7]. Protein painting employs small molecular dyes that covalently “paint” the surface of multi-protein complexes and block access to enzymatic (e.g., trypsin) digestion during MS analysis (Figure 1) [8]. Through paint coverage, this method allows for the identification of ligand-binding regions useful for drug development at the nAChR site. In addition, protein painting MS complements other experimental tools, including computational structural modeling and site-directed mutagenesis of the nAChR ligand binding pocket [9]. The protein paint technique can be easily adapted and optimized (Figure 2) on a case-by-case basis, allowing for its utility in various studies. The components of the protein paint assay are widely available and include inexpensive protein dyes, common chemical reagents, and MS instrumentation that are accessible at research institutions. Results from protein painting can guide structure-activity relationship (SAR) studies in lead compound development during drug screening and may allow for the identification of additional allosteric binding sites within the nAChR. Lastly, protein painting MS can also assess for the potential binding of new ligands to the nAChR when there is no prior knowledge of interactions.
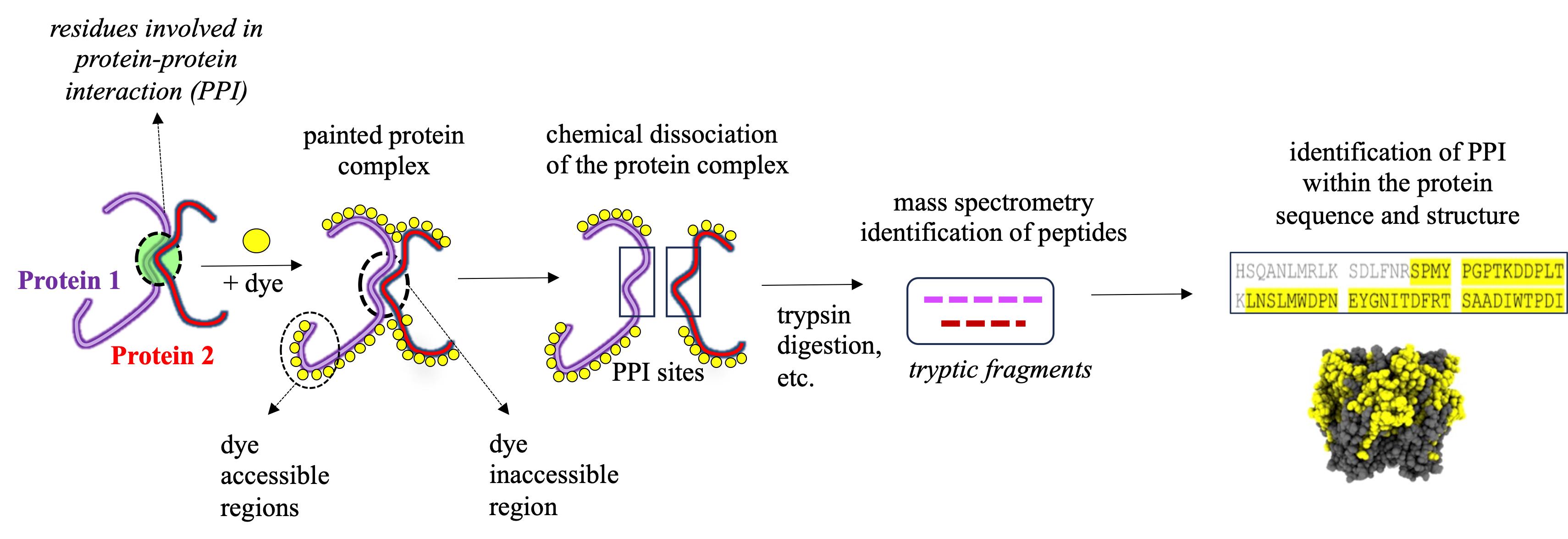
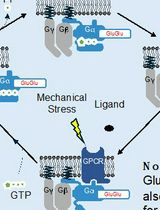
Figure 1. Identification of protein–protein interaction (PPI) sites through mass spectrometry (MS) protein painting. Protein painting utilizes the ability of various protein dyes to covalently bind to non-occupied (dye accessible) regions within a multi-protein complex. Dye inaccessible regions that correspond to possible PPI interaction sites are identified through MS analysis.
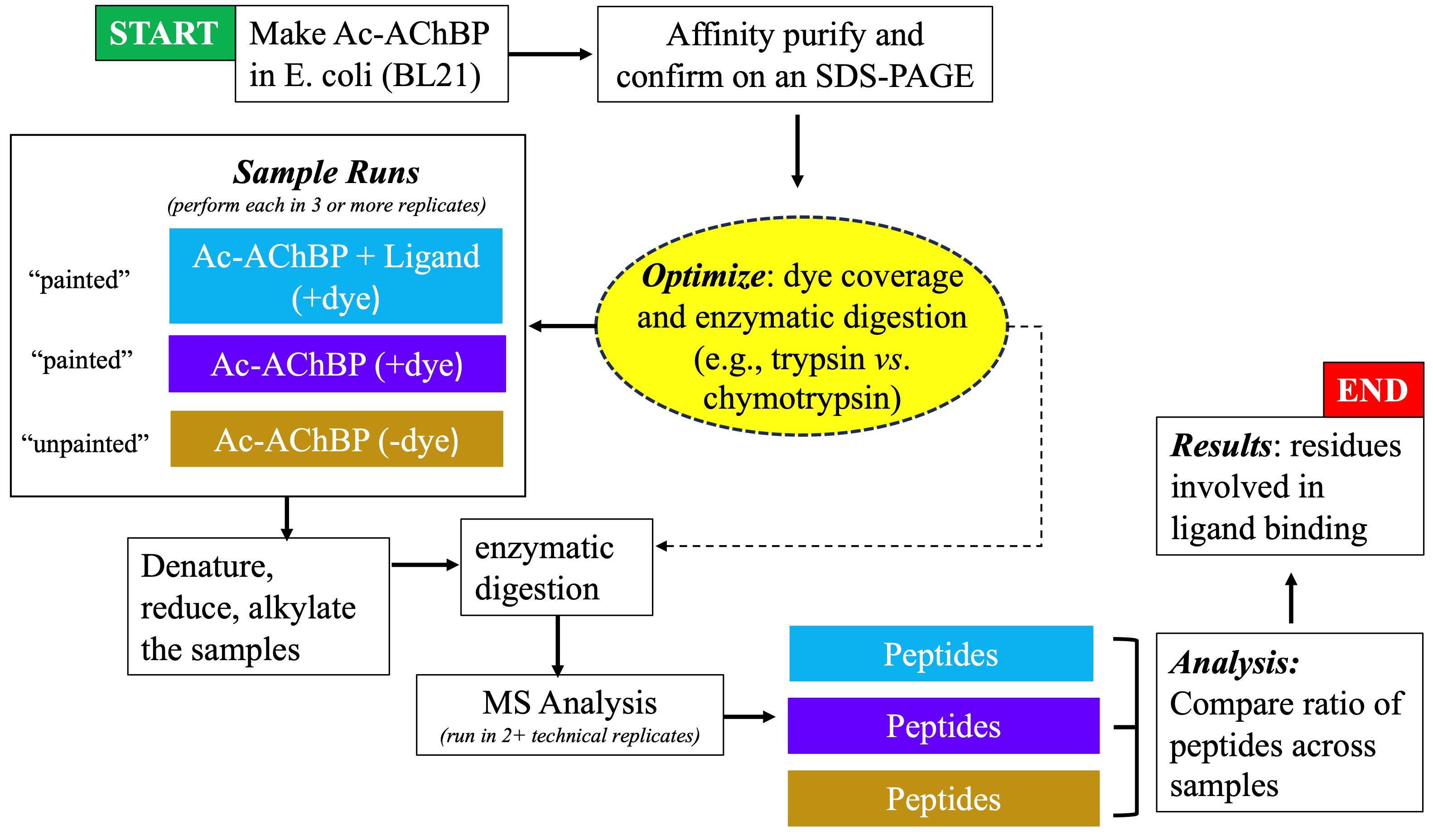
Figure 2. Flowchart showing the progression of experimental steps during the protein painting study
Materials and reagents
Biological materials
1. Escherichia coli BL21 (DE3) competent cells (Thermo Scientific, catalog number: EC0114)
Reagents
1. Invitrogen alpha-bungarotoxin conjugates (Thermo Fisher Scientific, catalog number: B1601)
2. Amyloid β-Protein 1-42 (Bachem, catalog number: 4014447)
3. 4-Nitrobenzenediazonium tetrafluoroborate (TCI, catalog number: N0137)
4. Disuccinimidyl suberate (DSS) (Thermo Fisher Scientific, catalog number: A39267)
5. Atto 425 NHS ester (Sigma-Aldrich, catalog number: 16805)
6. Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418)
7. Anhydrous DMSO (Thermo Fisher Scientific, catalog number: D12345)
8. Choline (Acros Organics, catalog number: 110290500)
9. Nicotine (Sigma-Aldrich, catalog number: N3876)
10. Lysozyme (Thermo Fischer Scientific, catalog number: 89833)
11. Sequencing-grade trypsin (Promega, catalog number: V5111)
12. Sequencing-grade chymotrypsin (Promega, catalog number: V1061)
13. DNase (Sigma-Aldrich, catalog number: DN25-1G)
14. Complete EDTA-free mini protease inhibitor (Thermo Fisher Scientific, catalog number: A32955)
15. Protein ladder (Thermo Fischer Scientific, catalog number: 26619)
16. Phosphate-buffered saline (PBS) (VWR, catalog number: 45001-130)
17. Dithiothreitol (DTT) (Thermo Fischer Scientific, catalog number: R0861)
18. Urea (Thermo Fischer Scientific, catalog number: AC424581000)
19. Iodoacetamide (Sigma-Aldrich, catalog number: I1149)
20. Ammonium bicarbonate (Thermo Fischer Scientific, catalog number: A643)
21. Acetic acid (Thermo Fischer Scientific, catalog number: A38-500)
22. Acetonitrile (ACN) (Fischer Scientific, catalog number: A21-1)
23. Trifluoroacetic acid (TFA) (Thermo Fischer Scientific, catalog number: A116-1AMP)
24. LB broth (Thermo Fischer Scientific, catalog number: BP1426-2)
25. Ampicillin (AMP) (Thermo Fischer Scientific, catalog number: 11593027)
26. Isopropyl β-d-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I6758)
27. Tris base (Thermo Fischer Scientific, catalog number: BP152)
28. Sodium chloride (NaCl) (Thermo Fischer Scientific, catalog number: S25541)
29. Glycerol (Thermo Fischer Scientific, catalog number: J61059.AP)
30. Imidazole (Thermo Fischer Scientific, catalog number: 03196-500)
31. Triton X-100 (Sigma-Aldrich, catalog number: X100)
32. Coomassie Brilliant Blue G 250 (Sigma-Aldrich, catalog number: 1.15444)
33. Methanol (VWR, catalog number: BDH1135-4LG)
Solutions
1. Lysis buffer (see Recipes)
2. Resuspension buffer (Buffer A) (see Recipes)
3. Elution buffer (Buffer B) (see Recipes)
4. Final buffer (see Recipes)
5. Diazo dye (see Recipes)
6. NHS ester dye (see Recipes)
7. Sample buffer (see Recipes)
8. Coomassie solution (see Recipes)
Recipes
Notes:
1. All buffers should be autoclaved or filtered before use and kept at 4 °C.
2. All dyes should be prepared fresh, immediately before use and be protected from light exposure as much as possible.
1. Lysis buffer
20 mM Tris, pH 8.0
150 mM NaCl
10% glycerol
2. Resuspension buffer (Buffer A)
20 mM Tris, pH 8.0
150 mM NaCl
10% glycerol
10 mM imidazole
1% Triton X-100
3. Elution buffer (Buffer B)
20 mM Tris, pH 8.0
150 mM NaCl
10% glycerol
250 mM imidazole
1% Triton X-100
4. Final buffer
20 mM Tris, pH 8.0
150 mM NaCl
10% glycerol
5. Diazo dye
Solid 4-nitrobenzenediazonium tetrafluoroborate
1× PBS
Make 5 mg/mL stock. For each painted sample, you need 5 μL.
6. NHS ester dye
1 mg/mL Atto 425 NHS ester
Anhydrous DMSO
Make 5 mg/mL stock. For each painted sample, you need 5 μL.
7. Sample buffer
20% ACN
2% TFA
Sterile dd/diH2O
8. Coomassie solution
50% methanol
10% acetic acid
0.5% Coomassie Brilliant Blue G 250
Sterile dd/diH2O
Laboratory supplies
1. Eppendorf protein Lo-Bind 1.5 mL tubes (Eppendorf, catalog number: 022431081)
2. Sephadex G25 spin columns (Cytiva, catalog number: 27532501)
3. PierceTM C-18 spin columns (Thermo Fisher Scientific, catalog number: 89873)
4. Ni-charged IMAC column (Bio-Rad, catalog number: 12009287)
5. 30 kDa Amicon ultra centrifugal filter (Sigma-Aldrich, catalog number: UFC903008)
6. NuPAGE 4%–12% Bis-Tris gradient gel (Thermo Fisher Scientific, catalog number: NP0322BOX)
7. Graduated Erlenmeyer flasks (Sigma-Aldrich, catalog number: CLS4980250 and CLS49802L)
Equipment
1. Centrifuge 5810R (Eppendorf, catalog number: 022625501)
2. Sorvall WX+ Ultracentrifuge (Thermo Fisher Scientific, catalog number: 75000100)
3. Water bath (Thermo Fisher Scientific, catalog number: 15-474-18)
4. Sonicator (Thermo Fisher Scientific, catalog number: 15-338-281)
5. Tube rotator (Thermo Fisher Scientific, catalog number: 13-687-12Q)
6. Spectrophotometer (Cole Parmer, catalog number: EW-83059-10)
7. Exploris Orbitrap 480 coupled with an EASY-nLC 1200 HLPC system (Thermo Fisher Scientific, catalog number: BRE725533)
8. Reverse-phase PepMap RSLC C18 LC column (Thermo Fisher Scientific, catalog number: 164534)
9. Milli-Q IQ 7003/05/10/15 water purification system (Millipore Sigma, catalog number: C205110)
10. Autoclave
11. Incubated and refrigerated console shakers (Thermo Fisher Scientific, catalog number: SHKE435HP)
Software and datasets
1. Proteome Discoverer v2.3 (Thermo Fisher Scientific, 09/27/2019)
2. UCSF ChimeraX v1.6 (05/09/2023) (https://www.cgl.ucsf.edu/chimerax/)
3. Microsoft Excel 2016
4. RCSB Protein Data Bank (RCSB PDB) (https://www.rcsb.org/)
5. NCBI database, open access
6. Prism v10.2.0 (GraphPad, 03/26/2024)
Procedure
文章信息
稿件历史记录
提交日期: Aug 21, 2024
接收日期: Nov 20, 2024
在线发布日期: Dec 5, 2024
出版日期: Jan 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Graur, A., Erickson, N. and Kabbani, N. (2025). Using Protein Painting Mass Spectrometry to Define Ligand Receptor Interaction Sites for Acetylcholine Binding Protein. Bio-protocol 15(2): e5163. DOI: 10.21769/BioProtoc.5163.
分类
神经科学 > 细胞机理 > 受体-配体结合
生物化学 > 蛋白质 > 相互作用 > 蛋白质-蛋白质相互作用
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link