- EN - English
- CN - 中文
Cloning a Chloroplast Genome in Saccharomyces cerevisiae and Escherichia coli
在酿酒酵母和大肠杆菌中克隆叶绿体基因组
(§ Technical contact) 发布: 2025年01月20日第15卷第2期 DOI: 10.21769/BioProtoc.5162 浏览次数: 4081
评审: Lucy XieSean L. BeckwithEmilia Krypotou

相关实验方案
I-PREFR:基于反向PCR的无酶单向策略,利用自杀载体在细菌中快速实现无标记染色体基因缺失与重建
Rekha Rana [...] Prabhu B. Patil
2025年05月20日 2343 阅读
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1231 阅读
Abstract
Chloroplast genomes present an alternative strategy for large-scale engineering of photosynthetic eukaryotes. Prior to our work, the chloroplast genomes of Chlamydomonas reinhardtii (204 kb) and Zea mays (140 kb) had been cloned using bacterial and yeast artificial chromosome (BAC/YAC) libraries, respectively. These methods lack design flexibility as they are reliant upon the random capture of genomic fragments during BAC/YAC library creation; additionally, both demonstrated a low efficiency (≤ 10%) for correct assembly of the genome in yeast. With this in mind, we sought to create a highly flexible and efficient approach for assembling the 117 kb chloroplast genome of Phaeodactylum tricornutum, a photosynthetic marine diatom. Our original article demonstrated a PCR-based approach for cloning the P. tricornutum chloroplast genome that had 90%–100% efficiency when screening as few as 10 yeast colonies following assembly. In this article, we will discuss this approach in greater depth as we believe this technique could be extrapolated to other species, particularly those with a similar chloroplast genome size and architecture.
Key features
• Large fragments of the chloroplast genome can be readily amplified through PCR from total algal DNA isolate.
• Assembly protocol can be completed within a day, and yeast colonies harboring chloroplast genomes can be obtained in as few as 4–5 days.
• Cloned genomes isolated from yeast transformants can be moved to Escherichia coli through electroporation.
Keywords: Synthetic biology (合成生物学)Graphical overview
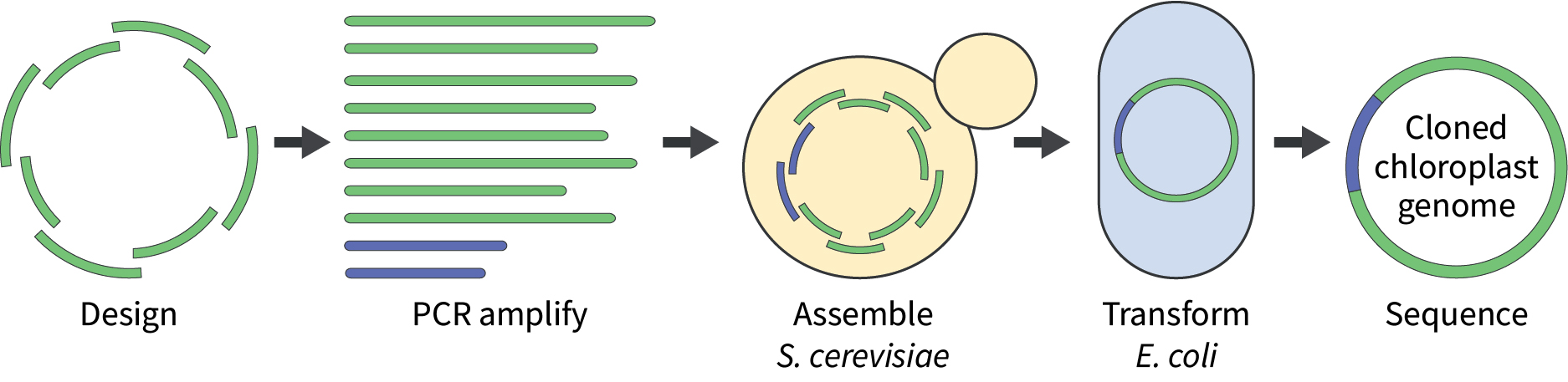
The chloroplast genome is split into overlapping fragments (green), which are then PCR-amplified along with a suitable cloning vector (blue). The fragments are transformed into Saccharomyces cerevisiae for assembly, where they will recombine to form the cloned chloroplast genome. DNA can be isolated from yeast transformants and electroporated into Escherichia coli for further analysis. Ultimately, to confirm the capture of the cloned genome, DNA from representative yeast and/or E. coli transformants is sequenced.
Background
The ability to assemble, deliver, and install whole synthetic genomes provides the utmost control when engineering an organism. However, synthesizing large fragments of DNA (i.e., > 10 kb) is still prohibitively expensive for most academic labs, whilst cloning and transforming partial or whole chromosomes remains technically challenging, if not unexplored, in most species. The model systems for whole-genome replacement have been limited to comparatively simple organisms with well-established DNA manipulation techniques (e.g., viruses [1,2], bacteria [3,4], and Saccharomyces cerevisiae [5]). Given the exciting possibilities that whole-genome replacement offers, there is a growing demand to establish other biological chassis capable of large-scale engineering feats [6]. Establishing a photosynthetic chassis is of particular interest to the biotechnology and bioeconomy sectors, as photosynthetic organisms can capture and use atmospheric CO2 as a carbon source, forgoing the need for energetically costly carbon inputs.
Photosynthetic eukaryotes possess distinct nuclear, chloroplast, and mitochondrial genomes. The aforementioned organelles typically contain multiple copies of a single, highly reduced chromosome, making these genomes more feasible to synthesize and “replace” compared to nuclear chromosomes, which are larger and more complex. The chloroplast genome is a particularly interesting target for genome replacement as the organelle serves as a hub for several cellular biosynthesis pathways of industrial relevance (e.g., starch, amino acid, and fatty acid synthesis). Despite this, there are only two published papers demonstrating the potential for cloning an entire chloroplast genome outside of the organelle. The first paper describes the capture of the Zea mays chloroplast genome (140 kb) in Saccharomyces cerevisiae [7]. Here, high molecular weight DNA was isolated from Z. mays, then shorn and ligated into a yeast artificial chromosome (YAC) backbone. More than 10,000 yeast clones were screened, with only one clone demonstrating what appeared to be the complete Z. mays chloroplast genome. The second paper describes the assembly of the Chlamydomonas reinhardtii chloroplast genome (204 kb) from six overlapping fragments, of which four had been previously captured in a bacterial artificial chromosome (BAC) library [8]. Here, 3 out of 30 yeast transformants contained the correctly assembled chloroplast genome following yeast assembly. Creating BAC/YAC libraries is technically demanding, time consuming, and does not permit the utmost design flexibility for assembling large constructs as it is dependent upon whichever fragments were generated during the library preparation stage. An efficient and tractable strategy for assembling whole chloroplast genomes is a necessary first step to achieve the full potential of this unique chassis.
We sought to design and test alternative assembly strategies for cloning whole chloroplast genomes. Phaeodactylum tricornutum was selected as our model organism due to its ease of propagation and rapidly growing toolbox for genetic engineering. Here, we present a strategy for cloning the P. tricornutum chloroplast genome that demonstrated 90%–100% efficiency when screening as few as 10 yeast colonies and 3 Escherichia coli colonies following whole-genome assembly and transformation, respectively. We believe this efficient and tractable method for cloning the P. tricornutum chloroplast genome could be extrapolated to other photosynthetic eukaryotes, particularly those with similar genome sizes and architectures (e.g., Thalassiosira pseudonana).
Materials and reagents
Biological materials
1. P. tricornutum liquid culture [Culture Collection of Algae, the University of Texas at Austin (UTEX), catalog number: 646]
2. E. coli TransforMax EPI300 cells (LGC Biosearch Technologies, Lucigen, catalog number: EC300110)
3. S. cerevisiae VL6-48 culture [American Type Culture Collection (ATCC), catalog number: MYA-3666]
Reagents
1. 1 kb DNA ladder (New England Biolabs, catalog number: N3232L)
2. 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
3. Adenine hemisulfate salt (Sigma-Aldrich, catalog number: A2545)
4. Agar A (Bio Basic, catalog number: FB0010)
5. Agarose (FroggaBio, catalog number: A87-500G)
6. Bacteriological peptone (BioShop, catalog number: PEP403)
7. Bacto agar (Becton Dickinson, catalog number: 214030)
8. Bio-tryptone (BioShop, catalog number: TRP402)
9. Boric acid (Bio Basic, catalog number: BB0044)
10. Calcium chloride, dihydrate (BioShop, catalog number: CCL302)
11. Cetyltrimethylammonium bromide (CTAB) (Sigma-Aldrich, catalog number: H6269)
12. Chloramphenicol (Bio Basic, catalog number: CB0118)
13. Chloroform:isoamyl alcohol, 24:1 (Bio Basic, catalog number: CB0351)
14. Cobalt (II) sulfate, heptahydrate (Sigma-Aldrich, catalog number: CDS004010)
15. Complete media glucose broth minus tryptophan (Teknova, catalog number: C7131)
Note: This product is no longer available; as an alternative, we suggest minimal SD base (Takara, catalog number: 630411) and -Trp DO supplement (Takara, catalog number: 630413).
16. Complete media glucose broth minus histidine and uracil (Teknova, catalog number: C7221)
Note: This product is no longer available; as an alternative, we suggest minimal SD base (Takara, catalog number: 630411) in addition to the minus histidine and uracil (-His/-Ura) DO supplement (Takara, catalog number: 630422).
17. Copper (II) sulfate, pentahydrate (Bio Basic, catalog number: CDB0063)
18. Cyanocobalamin, i.e., Vitamin B12 (BioShop, catalog number: VIT271)
19. D-biotin (Bio Basic, catalog number: BB0078)
20. D-glucose (BioShop, catalog number: GLU501)
21. D-sorbitol (BioShop, catalog number: SOR508)
22. DNA gel loading dye (New England Biolabs, catalog number: B7024S)
23. DpnI restriction enzyme (New England Biolabs, catalog number: R0176)
24. Ethylenediaminetetraacetic acid disodium salt, dihydrate (EDTA) (Bio Basic, catalog number: EB0185)
25. Ethanol, 95% purity (Greenfield Global, catalog number: P016EA95)
26. Ethanol, anhydrous (Greenfield Global, catalog number: P006EAAN)
27. Ethidium bromide (BioShop, catalog number: ETB444)
28. Glycerol (Bio Basic, catalog number: GB0232)
29. Iron chloride, hexahydrate (Bio Basic, catalog number: FD0201)
30. Isopropanol, min. 99.5% purity (Bioshop, catalog number: SO920)
31. L-arabinose (BioShop, catalog number: ARB222)
32. Lysozyme, egg white (BioShop, catalog number: LYS702)
33. Magnesium chloride, hexahydrate (BioShop, catalog number: MAG510)
34. Manganese (II) chloride, tetrahydrate (Sigma-Aldrich, catalog number: 805930)
35. Molybdic acid, sodium salt (Bio Basic, catalog number: MB0358)
36. Nickel (II) sulfate, hexahydrate (BioShop, catalog number: NIC700)
37. Phenol:chloroform:isoamyl alcohol, 25:24:1 (Fisher Scientific, catalog number: 15593031)
38. Polyethylene glycol 8000 (Fisher Scientific, catalog number: BP233-1)
39. Potassium acetate (Bio Basic, catalog number: PRB0438)
40. Potassium bromide (BioShop, catalog number: POB333)
41. Potassium chloride (Bio Basic, catalog number: PB0440)
42. Potassium chromate (Thermo Scientific Chemicals, catalog number: AC447201000)
43. Proteinase K solution (BioShop, catalog number: PRK222)
44. RNase A (QIAGEN, catalog number: 19101)
45. Selenious acid (Thermo Scientific Chemicals, catalog number: 211176)
46. Sodium acetate (Bio Basic, catalog number: SB1611)
47. Sodium bicarbonate (Bio Basic, catalog number: SB0482)
48. Sodium chloride (BioShop, catalog number: SOD004)
49. Sodium dodecyl sulfate (BioShop, catalog number: SDS001)
50. Sodium hydroxide (BioShop, catalog number: SHY700)
51. Sodium fluoride (BioShop, catalog number: SFL001)
52. Sodium nitrate (Bio Basic, catalog number: SD0484)
53. Sodium orthovanadate (BioShop, catalog number: SOV850)
54. Sodium phosphate, dibasic (BioShop, catalog number: SPD307)
55. Sodium phosphate, monobasic (BioShop, catalog number: SPM400)
56. Sodium sulfate (BioShop, catalog number: SOS513)
57. Thiamine hydrochloride, i.e., Vitamin B1 (Sigma-Aldrich, catalog number: T4625)
58. Tris hydrochloride (Bio Basic, catalog number: TB0103)
59. Yeast extract (BioShop, catalog number: YEX555)
60. Zinc sulfate, heptahydrate (Bio Basic, catalog number: ZB2906)
61. Zymolyase, 20,000 units/g (BioShop, catalog number: ZYM001)
Solutions
1. Biotin stock solution, 0.1% (w/v)
2. Boric acid stock solution, 0.1% (w/v)
3. Chilled isopropanol, 100% (v/v), -20 °C
4. Chilled ethanol, 70% (v/v), -20 °C
5. Cobalt (II) sulfate stock solution, 1% (w/v)
6. Copper (II) sulfate stock solution, 0.98% (w/v)
7. Cyanocobalamin stock solution, 0.1% (w/v)
8. D-sorbitol, 1 M
9. Ethylenediaminetetraacetic acid solution, 0.5 M, pH 8.0 (EDTA)
10. Glucose, 1 M
11. Glycerol, 50% (v/v)
12. Magnesium chloride, 1 M
13. Manganese (II) chloride stock solution, 18% (w/v)
14. Molybdic acid stock solution, 0.63% (w/v)
15. Nickel (II) sulfate stock solution, 0.27% (w/v)
16. Potassium chloride, 250 mM
17. Potassium chromate stock solution, 0.194% (w/v)
18. Selenious acid stock solution, 0.13% (w/v)
19. Sodium acetate, 3 M, pH 5.2
20. Sodium fluoride stock solution, 0.1% (w/v)
21. Sodium hydroxide solution, 1 M
22. Sodium orthovanadate stock solution, 0.184% (w/v)
23. Synthetic media lacking -Trp
24. Synthetic media lacking -His/Ura
25. Tris hydrochloride, 1 M, pH 8.0 (Tris-HCl)
26. Zinc sulfate stock solution, 2.2% (w/v)
DNA extraction
1. Buffer P1 (see Recipes)
2. Buffer P2 (see Recipes)
3. Buffer P3 (see Recipes)
4. CTAB lysis buffer (see Recipes)
Culturing E. coli
1. LB broth (see Recipes)
2. SOB broth (see Recipes)
3. SOC broth (see Recipes)
Culturing yeast
1. 2× YPAD broth (see Recipes)
2. Bacto-agar, 2% (w/v) (see Recipes)
3. Complete media (CM) glucose broth minus histidine and uracil (see Recipes)
Yeast assembly
1. SPEM solution (see Recipes)
2. Zymolase solution (see Recipes)
3. STC solution (see Recipes)
4. PEG-8000 solution (see Recipes)
5. SOS media (see Recipes)
Culturing P. tricornutum
1. NP stock, 500× (see Recipes)
2. L1 trace metals stock, 1,000× (see Recipes)
3. F/2 vitamin stock solution, 2,000× (see Recipes)
4. Anhydrous salts solution, 2× (see Recipes)
5. Hydrous salts solution, 2× (see Recipes)
6. L1 media (see Recipes)
Recipes
A. DNA extraction
1. Buffer P1
Store at 4 °C for < 12 months. Recipe made publicly available by QIAGEN.
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl (1 M, pH 8.0) | 5.0 × 10-2 M | 5 mL |
| EDTA (0.5 M, pH 8.0) | 1.0 × 10-2 M | 2 mL |
| RNAse A (100 mg/mL) | 100 μg/mL | 100 μL |
| ddH2O | n/a | 92.9 mL |
| Total | n/a | 100 mL |
2. Buffer P2
Store at room temperature for < 24 months. Recipe made publicly available by QIAGEN.
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium hydroxide | 2.0 × 10-1 M | 4 g |
| Sodium dodecyl sulfate | 1% (w/v) | 5 g |
| ddH2O | n/a | Top up to 500 mL |
| Total | n/a | 500 mL |
3. Buffer P3
Store at room temperature for < 24 months. Recipe made publicly available by QIAGEN.
| Reagent | Final concentration | Amount |
|---|---|---|
| Potassium acetate | 3 M | 147.2 g |
| ddH2O | n/a | Top up to 500 mL |
| Total | n/a | 500 mL |
4. CTAB lysis buffer
Store at 4 °C for < 12 months. Recipe was obtained from Giguere et al. [9].
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium chloride | 1.4 M | 4.1 g |
| Tris-HCl (1 M, pH 8.0) | 2.0 × 10-1 M | 10 mL |
| EDTA (0.5 M, pH 8.0) | 5.0 × 10-2 M | 5 mL |
| CTAB | 2% (w/v) | 1 g |
| RNAse A (100 mg/mL) | 250 μg/mL | 125 μL |
| ddH2O | n/a | Top up to 50 mL |
| Total | n/a | 50 mL |
B. Culturing E. coli
1. LB broth
Sterilize by autoclaving, then store at room temperature for < 12 months. To make solid media, add 1.5 g of agar A for every 100 mL of LB (1.5%, w/v) prior to autoclaving.
| Reagent | Final concentration | Amount |
|---|---|---|
| Bio-tryptone | n/a | 10 g |
| Sodium chloride | 1.71 × 10-1 M | 10 g |
| Yeast extract | n/a | 5 g |
| dH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
2. SOB broth
Mix all the components, excluding the magnesium chloride, then adjust the pH to 7.0 using NaOH and/or HCl as necessary. Sterilize by autoclaving and allow to cool to room temperature. Then, aseptically add 10 mL of sterile magnesium chloride solution. The magnesium chloride solution can be sterilized by autoclaving as well. SOB broth can be stored at room temperature for < 12 months.
| Reagent | Final concentration | Amount |
|---|---|---|
| Bio-tryptone | n/a | 20 g |
| Yeast extract | n/a | 5 g |
| Sodium chloride | 1.0 × 10-2 M | 0.5 g |
| Potassium chloride, 250 mM | 2.5 × 10-3 M | 10 mL |
| dH2O | n/a | Top up to 990 mL* |
| Magnesium chloride, 1 M | 1.0 × 10-3 M | 10 mL |
| Total | n/a | 1,000 mL |
3. SOC broth
Aseptically add 20% (w/v) filter-sterilized glucose to the sterile SOB broth. We recommend preparing 50 mL sterile aliquots of this media as it is very susceptible to contamination.
| Reagent | Final concentration | Amount |
|---|---|---|
| SOB broth | n/a | 1,000 mL |
| Glucose, 1 M | 2.0 × 10-2 M | 20 mL |
| Total | n/a | 1,020 mL |
C. Culturing yeast
1. 2× YPAD broth
Filter sterilize into a sterile glass bottle, then store at room temperature for < 12 months. D-glucose (i.e., dextrose) can burn if autoclaved, which will impact the growth of yeast. To make solid media, combine 2× YPAD with melted 2% (w/v) bacto-agar. Ensure the YPAD and bacto-agar have been equilibrated to 60 °C before combining in a 1:1 ratio to make 1% bacto-agar 1× YPAD plates. If using pre-mixed YPD agar (e.g., Sigma-Aldrich, catalog number: Y1500), combine 130 g/L of powder with dH2O and autoclave for 15 min at 121 °C to sterilize.
| Reagent | Final concentration | Amount |
|---|---|---|
| Yeast extract | n/a | 20 g |
| Bacteriological peptone | n/a | 40 g |
| D-glucose | 2.2 × 10-1 M | 40 g |
| Adenine hemisulfate | n/a | 160 mg |
| dH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
2. Bacto-agar, 2% (w/v)
Autoclave to sterilize, ensuring that the glass bottle is no more than 80% full to avoid boiling over. Store at room temperature for < 24 months. The recipe below lists the volume we would prepare in a 500 mL glass bottle.
| Reagent | Final concentration | Amount |
|---|---|---|
| Bacto-agar | 2% (w/v) | 8 g |
| dH2O | n/a | Top up to 400 mL |
| Total | n/a | 400 mL |
3. Complete media (CM) glucose broth minus histidine and uracil
Adjust pH to 6.0, then autoclave to sterilize; store at room temperature for < 12 months. Only include D-sorbitol if plating spheroplasted yeast (i.e., during yeast assembly); omit when plating yeast with intact cell walls. To make solid media, add 2 g of Bacto-agar for every 100 mL of media (2%, w/v). Recipe is based on the manufacturer’s guidelines (Teknova) and Karas et al. [10].
| Reagent | Final concentration | Amount |
|---|---|---|
| CM glucose broth minus histidine and uracil | n/a | 28.4 g |
| D-sorbitol | 1.0 M | 182 g |
| Adenine hemisulfate | 8.69 × 10-4 M | 160 mg |
| dH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
D. Yeast assembly
1. SPEM solution
Filter sterilize into a sterile glass bottle, then store at room temperature for < 12 months. Recipe was obtained from Karas et al. [10].
| Reagent | Final concentration | Amount |
|---|---|---|
| D-sorbitol | 1.0 M | 182 g |
| EDTA (0.5 M, pH 8.0) | 1.0 × 10-2 M | 20 mL |
| Sodium phosphate, dibasic | 1.47 × 10-2 M | 2.08 g |
| Sodium phosphate, monobasic | 2.32 × 10-3 M | 0.32 g |
| dH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
2. Zymolyase solution
Sterilize using a syringe filter, then store in sterile 1.5 mL tubes at -20 °C for < 12 months. The efficiency of zymolyase noticeably drops with every freeze-thaw event, so we suggest storing aliquots of 45–90 μL, which is enough for 1–2 assembly reactions. Recipe was obtained from Karas et al. [10].
| Reagent | Final concentration | Amount |
|---|---|---|
| Zymolyase | 400 units/mL | 200 mg |
| Tris-HCl (1 M, pH 8.0) | 1.0 × 10-1 M | 1 mL |
| Glycerol (50% v/v) | 25% (v/v) | 10 mL |
| ddH2O | n/a | 9 mL |
| Total | n/a | 20 mL |
3. STC solution
Filter sterilize into a sterile glass bottle, then store at room temperature for < 12 months. It is optimal to store this solution as 15 mL aliquots in sterile conical tubes. Recipe was obtained from Karas et al. [10].
| Reagent | Final concentration | Amount |
|---|---|---|
| D-sorbitol | 1.0 M | 182 g |
| Tris-HCl (1 M, pH 8.0) | 1.0 × 10-2 M | 10 mL |
| Calcium chloride, dihydrate | 1.0 × 10-2 M | 1.47 g |
| Magnesium chloride (1 M) | 2.5 × 10-3 M | 2.5 mL |
| dH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
4. PEG-8000 solution
Adjust pH to 8.0 with sodium hydroxide solution, then filter sterilize into a sterile glass bottle; store at 4 °C for < 12 months. PEG-8000 will depolymerize over time, thereby becoming less effective for yeast assembly. This process is exacerbated if the solution is left at room temperature. It is important to check the pH of this solution ahead of its use in assembly; when stored correctly, the pH should remain at or near 8.0 for up to a year. If the pH drops below this, dispose of the solution and remake it. Recipe was obtained from Karas et al. [10].
| Reagent | Final concentration | Amount |
|---|---|---|
| PEG-8000 | 20% (w/v) | 20 g |
| Calcium chloride, dihydrate | 1.0 × 10-2 M | 0.147 g |
| Magnesium chloride (1 M) | 2.5 × 10-3 M | 250 μL |
| Tris-HCl (1 M, pH 8.0) | 1.0 × 10-2 M | 1 mL |
| dH2O | n/a | Top up to 100 mL |
| Total | n/a | 100 mL |
5. SOS media
Filter sterilize into a sterile glass bottle, then store at room temperature for < 12 months. It is optimal to store this solution as 15 mL aliquots in sterile conical tubes. Recipe was obtained from Karas et al. [10].
| Reagent | Final concentration | Amount |
|---|---|---|
| D-sorbitol | 1.0 M | 182 g |
| Bacteriological peptone | n/a | 5 g |
| Yeast extract | n/a | 2.5 g |
| Calcium chloride, dihydrate | 6.04 × 10-3 M | 0.888 g |
| dH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
E. Culturing P. tricornutum
1. NP stock, 500×
Filter sterilize into a sterile glass bottle, then store at room temperature for < 24 months. Recipe was obtained from Karas et al. [11].
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium nitrate | 4.4 M | 37.5 g |
| Sodium phosphate, monobasic | 1.80 × 10-1 M | 2.5 g |
| ddH2O | n/a | Top up to 100 mL |
| Total | n/a | 100 mL |
2. L1 trace metals stock, 1,000×
Filter sterilize into a sterile glass bottle, then store at 4 °C for < 24 months. Recipe was obtained from Karas et al. [11].
| Reagent | Final concentration | Amount |
|---|---|---|
| Iron chloride, hexahydrate | 1.17 × 10-2 M | 3.15 g |
| EDTA disodium salt, dihydrate | 1.17 × 10-2 M | 4.36 g |
| Copper (II) sulfate stock solution, 0.98% (w/v) | 9.81 × 10-6 M | 0.25 mL |
| Molybdic acid stock solution, 0.63% (w/v) | 7.81 × 10-5 M | 3.0 mL |
| Zinc sulfate stock solution, 2.2% (w/v) | 7.65 × 10-5 M | 1.0 mL |
| Cobalt (II) sulfate stock solution 1% (w/v) | 3.56 × 10-5 M | 1.0 mL |
| Manganese (II) chloride stock solution, 18% (w/v) | 9.10 × 10-4 M | 1.0 mL |
| Selenious acid stock solution, 0.13% (w/v) | 1.01 × 10-5 M | 1.0 mL |
| Nickel (II) sulfate stock solution, 0.27% (w/v) | 1.03 × 10-5 M | 1.0 mL |
| Sodium orthovanadate stock solution, 0.184% (w/v) | 1.00 × 10-5 M | 1.0 mL |
| Potassium chromate stock solution, 0.194% (w/v) | 9.99 × 10-6 M | 1.0 mL |
| ddH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
3. F/2 vitamin stock solution, 2,000×
Filter sterilize into a sterile glass bottle, then store at 4 °C for < 24 months. Recipe was obtained from Karas et al. [11].
| Reagent | Final concentration | Amount |
|---|---|---|
| Thiamine hydrochloride | 5.93 × 10-4 M | 200 mg |
| Biotin stock solution, 0.1% (w/v) | 4.09 × 10-5 M | 10 mL |
| Cyanocobalamin stock solution, 0.1% (w/v) | 7.38 × 10-7 M | 1 mL |
| ddH2O | n/a | Top up to 1,000 mL |
| Total | n/a | 1,000 mL |
4. Anhydrous salts solution, 2×
We use this immediately for making L1 media. If necessary, it can be filter sterilized and stored at 4 °C for < 12 months. Recipe was obtained from Karas et al. [11].
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium chloride | 8.38 × 10-1 M | 24.5 g |
| Sodium sulfate | 5.76 × 10-2 M | 4.09 g |
| Potassium chloride | 1.88 × 10-2 M | 0.7 g |
| Sodium bicarbonate | 4.76 × 10-3 M | 0.2 g |
| Potassium bromide | 1.68 × 10-3 M | 0.1 g |
| Boric acid stock solution, 0.1% (w/v) | 9.70 × 10-4 M | 3 mL |
| Sodium fluoride stock solution, 0.1% (w/v) | 1.43 × 10-4 M | 300 μL |
| ddH2O | n/a | Top up to 500 mL |
| Total | n/a | 500 mL |
5. Hydrous salts solution, 2×
We use this immediately for making L1 media. If necessary, it can be filter sterilized and stored at 4 °C for < 12 months. Recipe was obtained from Karas et al. [11].
| Reagent | Final concentration | Amount |
|---|---|---|
| Magnesium chloride, hexahydrate | 1.09 × 10-1 M | 11.1 g |
| Calcium chloride, dihydrate | 2.10 × 10-2 M | 1.54 g |
| ddH2O | n/a | Top up to 500 mL |
| Total | n/a | 500 mL |
6. L1 media
Adjust pH to 8.0 with sodium hydroxide solution, then filter sterilize into a sterile glass bottle; store at 4 °C for < 12 months. Note that this media lacks supplemental silica (sodium metasilicate nonahydrate) as it is not necessary for P. tricornutum growth. Recipe was obtained from Karas et al. [11].
| Reagent | Final concentration | Amount |
|---|---|---|
| Anhydrous salts solution, 2× | 1× | 500 mL |
| Hydrous salts solution, 2× | 1× | 500 mL |
| NP stock solution, 500× | 1× | 2 mL |
| L1 trace metals solution, 1,000× | 1× | 1 mL |
| F/2 vitamin solution, 2,000× | 1× | 0.5 mL |
| Total | n/a | ~1,000 mL |
Laboratory supplies
1. 0.2 mL PCR 8-strip tubes (FroggaBio, catalog number: STF-A120-S)
2. 1.5 mL tubes (FroggaBio, catalog number: 1210-001)
3. 15 mL conical tubes (FroggaBio, catalog number: TB15-500)
4. 50 mL conical tubes (FroggaBio, catalog number: TB50-500)
5. Bottle-top filters (≥ 500 mL capacity) with 0.2 μm PES membrane (Thermo Scientific, catalog number: 09-741-07)
6. Disposable hemocytometer, Neubauer-improved chamber (Fisher Scientific, SKC, catalog number: 22-600-100)
7. Disposable plastic cuvettes (VWR, catalog number: 97000-586)
8. Electrocuvettes, 2 mm (Fisher Scientific, catalog number: FB102)
9. Erlenmeyer flasks of various sizes (e.g., 100 mL, 250 mL)
10. EZ-10 Spin Column Plasmid DNA Miniprep kit (Bio Basic, catalog number: BS614)
11. EZ-10 Spin Column PCR Products Purification Kit (Bio Basic, catalog number: BS664)
12. Large-construct kit (QIAGEN, catalog number: 12462)
13. Liquid nitrogen (available at the institution)
14. Multiplex PCR kit (QIAGEN, catalog number: 206143)
15. Petri dishes, 100 × 15 mm (VWR, catalog number: 25384-088)
16. Porcelain mortar and pestle (Fisher Scientific, catalog numbers: FB961A and FB961K)
17. PrimeSTAR GXL DNA Polymerase kit (Takara, catalog number: R050A)
18. Two-sided disposable polystyrene cuvettes, 1.5–3.0 mL volume (VWR, catalog number: 97000-586)
19. Various sizes of glass bottles that can be sterilized
20. Pipette tips: 2 μL, 200 μL, 1,000 μL
Equipment
1. Biosafety cabinet (NuAire, model: LabGard ES NU-540)
2. Centrifuge with 15–50 mL tube capacity (e.g., Eppendorf, model: 5810, catalog number: 05-413-332)
3. Cryogenic dewar for liquid nitrogen
4. DeNovix spectrophotometer (DeNovix, model: DS-11, catalog number: DS-11)
5. Gel documentation system (Bio-Rad, model: ChemiDoc Imaging System, catalog number: 12003153)
6. Gel electrophoresis systems (Fisher Scientific, models: Owl EasyCast B1, B2 and B3, discontinued)
7. Gel power system (Fisher Scientific, catalog number: FB300Q)
8. Meker burner (Flinn Scientific, catalog number: AP1021)
9. Microcentrifuge with 1.5 mL tube capacity (Eppendorf, model: 5415C, discontinued)
10. pH probe (Sartorius, model: pHBasic+, discontinued)
11. Pipettes: P2, P20, P100, and P1000
12. Room or chamber at 18 °C with cool white light (i.e., blue-shifted) set to an intensity of 50–65 PPFD
13. Stationary/shaking incubators that can be set to 37 °C and 30 °C (Benchmark Scientific, catalog number: H1001-M)
14. Stir plate (Benchmark Scientific, catalog number: H4000-HS)
15. T100 thermal cycler (Bio-Rad, catalog number: 1861096)
Software and datasets
1. Benchling; free use, web-based platform (https://www.benchling.com/)
2. Image Lab; free use, application from Bio-Rad (version 6.1.0 build 7, standard edition)
3. Primer3web; free use, web-based platform (https://primer3.ut.ee/)
4. Sequenced and annotated P. tricornutum chloroplast genome; GenBank accession number EF067920.1, created by Oudot-Le Secq et al. [12]
Procedure
文章信息
稿件历史记录
提交日期: Aug 30, 2024
接收日期: Nov 7, 2024
在线发布日期: Nov 27, 2024
出版日期: Jan 20, 2025
版权信息
© 2025 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Walker, E. J. L. and Karas, B. J. (2025). Cloning a Chloroplast Genome in Saccharomyces cerevisiae and Escherichia coli. Bio-protocol 15(2): e5162. DOI: 10.21769/BioProtoc.5162.
Walker, E. J. L., Pampuch, M., Chang, N., Cochrane, R. R. and Karas, B. J. (2024). Design and assembly of the 117-kb Phaeodactylum tricornutum chloroplast genome. Plant Physiol. 194(4): 2217–2228.
分类
生物工程 > 合成生物学 > 基因修饰
分子生物学 > DNA > 染色体工程
微生物学 > 异源表达系统 > 酿酒酵母
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link