- EN - English
- CN - 中文
CRISPR/Cas9-Based Protocol for Precise Genome Editing in Induced Pluripotent Stem Cells
基于CRISPR/Cas9的诱导多能干细胞精确基因组编辑方案
发布: 2024年12月20日第14卷第24期 DOI: 10.21769/BioProtoc.5141 浏览次数: 2931
评审: Samantha HallerPhilipp WörsdörferRakesh Bam
Abstract
The advent of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-based genome editing has marked a significant advancement in genetic engineering technology. However, the editing of induced pluripotent stem cells (iPSCs) with CRISPR presents notable challenges in ensuring cell survival and achieving high editing efficiency. These challenges become even more complex when considering the specific target site. P53 activation as a result of traditional CRISPR editing can lead to apoptosis, potentially worsening cell health or even resulting in cell death. Mitigating this apoptotic response can enhance cell survival post-CRISPR editing, which will ultimately increase editing efficiency. In our study, we observed that combining p53 inhibition with pro-survival small molecules yields a homologous recombination rate of over 90% when using CRISPR in human iPSCs. This protocol significantly streamlines the editing process and reduces the time and resources necessary for creating isogenic lines.
Key features
• The combination of p53 inhibition and pro-survival small molecules promotes cell survival and increases the efficiency of genome editing.
• Genome editing can be completed in as little as 8 weeks for iPSCs, significantly reducing the total time required.
• Achieves a homologous recombination rate of over 90% in human iPSCs.
Keywords: iPSCs (iPSCs)Graphical overview
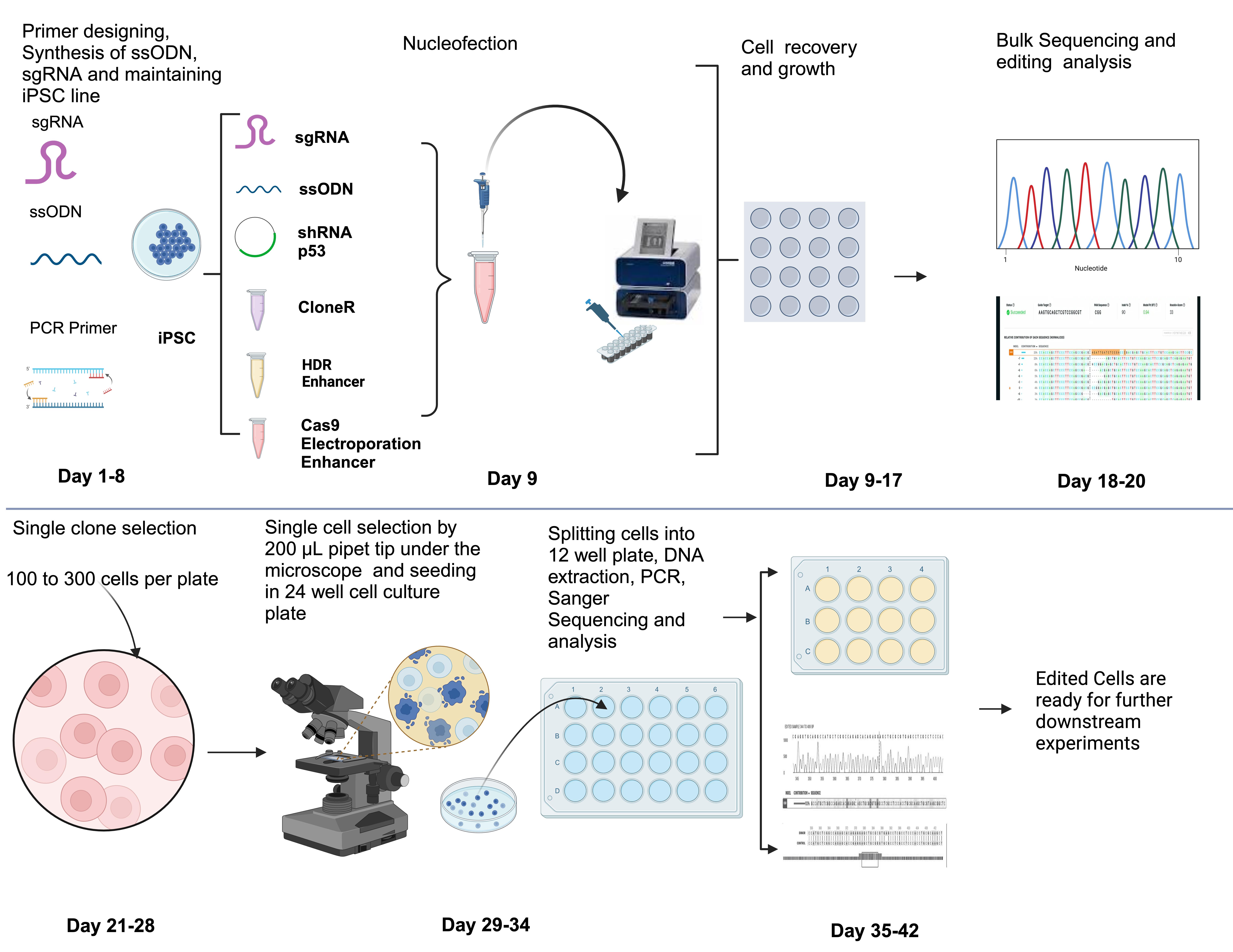
Outline of Protocol
Background
Genome editing has revolutionized biotechnology by allowing scientists to make precise changes to an organism's DNA. The ability to modify an organism's genome allows experts to study difficult evolutionary and medical problems in more complex systems [1]. The application of gene editing in induced pluripotent stem cells (iPSCs) to create genetically modified isogenic lines can enhance our understanding of how genetic mutations cause disease. Gene editing ranges from conceptually straightforward gene knockouts to more intricate modifications such as point mutations or whole gene insertions. However, currently, gene editing is still confronted with challenges [2–4]. In plant and animal models, clustered interspaced short palindromic repeats (CRISPR) have emerged as the tool of choice for genome editing [5]. Nowadays, the nuclease versions Cas9 and Cas12a are commonly used [1]. The CRISPR-Cas9 and Cas12a systems work with a guide RNA (gRNA) that is divided into two parts: a trans-activating CRISPR RNA (tracrRNA) that binds directly to the Cas nuclease to create ribonucleoprotein (RNP) and a CRISPR RNA (crRNA) that indicates the genomic target location [6]. When compared with previous editing techniques, the CRISPR-Cas innovation offers more precise targeting and editing proficiency as well as being easier and less expensive to configure, allowing it to be used more extensively [7–9]. However, the CRISPR-Cas editing systems still require improvement, particularly for point mutations. In certain cases, there may be several risk variants associated with a target gene, necessitating the creation of multiple lines with distinct single nucleotide polymorphisms (SNPs) or the sequential insertion of genetic modifications. Therefore, to enable more laboratories to produce the needed genetically modified lines, it is essential to develop a gene editing technique that is both highly efficient and easily adaptable. Currently, our attention is on overcoming the barriers to introducing point mutation in cell lines and improving the method to be as effective as possible. Editing efficiency may be increased by preventing cell death with the use of a Rho-related protein kinase (ROCK) inhibitor and blocking the p53 pathway [7]. In this protocol, we used pro-survival small molecules in conjunction with p53 inhibition to attain a homologous recombination rate exceeding 90%.
Materials and reagents
Biological materials
iPSC line (human NDC1) [10]
Reagents
mTeSR Plus basal (Stem Cell Technologies, catalog number: 100-0274)
mTeSR Plus 5× supplement (Stem Cell Technologies, catalog number: 100-0275)
CloneR (Stem Cell Technologies, catalog number: 05888 10 mL)
Revitacell supplement (Gibco, catalog number: A2644501 100×)
1× D-PBS (Gibco, catalog number: 10010049 500 mL)
DMEM/F12 (Gibco, catalog number: A4192001 500 mL)
Matrigel (Corning, catalog number: 47743-706 5 mL)
Accutase (VWR, catalog number: AT104 100 mL)
ReLeSR (Stem Cell Technologies, catalog number: 100-0484)
Alt-R Cas9 HDR enhancer (stock solution 3 mM) (IDT, catalog number: 1081062)
pmaxGFP (LONZA, catalog number: V4XP3032 50 μL at 1 μg/μL)
P3 primary cell nucleofector solution (LONZA, catalog number: V4XP3032 675 μL)
P3 primary cell nucleofector supplement (LONZA, catalog number: V4XP3032 150 μL)
Single-guide RNA (sgRNA) (IDT custom-made, 100 μM)
Alt-R S.p. HiFi Cas9 Nuclease V3 (IDT, catalog number: 10810559 500 μg at 10 μg/μL)
Single-strand oligo donor (ssODN) (IDT, 10 nmol)
shp53-f2 plasmid (Addgene)
Alt-R Cas9 electroporation enhancer (IDT, catalog number: 1075915 100 μm)
Molecular-grade H2O (Cytiva, catalog number: 82007-334 500 mL)
Zymo quick DNATM MicroPrep kit (Zymo Research, catalog number: D3021)
Agarose (VWR Life Sciences, catalog number: N605-500G)
Zymo CleanTM Gel DNA Recovery kit (Zymo Research, catalog number: D4002)
Solutions
Ribonucleoprotein (RNP) complex (see Recipes)
CloneR media (see Recipes)
Nucleofection reaction solution (see Recipes)
Nucleofection media (see Recipes)
P3 nucleofection solution mix (see Recipes)
mTeSR complete media (see Recipes)
Matrigel (see Recipes)
Recipes
RNP complex
This should be freshly prepared.
Reagent Final concentration Volume Cas9 nuclease V3 64 mM 0.8 μL sgRNA 100 µM 1.2 μL D-PBS (1×) 3.0 μL Total 5.0 μL CloneR media
This should be freshly prepared. Can be stored at 4 °C for one week.
Media component Volume mTeSR complete 8.9 mL Clone R 1 mL Revitacell 100 µL Total 10 mL Nucleofection reaction solution
This should be freshly prepared.
Component Test reaction GFP control No pulse control P3 primary cell nucleofector solution 1 μL 7 μL 1 μL shp53-f2 plasmid 1 μL 1 μL 1 μL Alt-R electroporation enhancer (100 μM) 1 μL 1 μL 1 μL ssODN (single-stranded oligodeoxynucleotide) (100 μM) 1 μL 0 μL 1 μL RNP complex 5 μL 0 μL 5 μL GFP (0.5 μg/μL) 0 μL 1 μL 0 μL Cells (1 million/reaction) (μL can be altered) 11 μL 11 μL 11 μL Total 20 μL 20 μL 20 μL Nucleofection media
This should be freshly prepared.
Component Volume CloneR media 1 mL Alt-R Cas9 HDR enhancer 10 µL P3 nucleofection solution mix
This should be freshly prepared.
Component Volume P3 primary cell nucleofector solution 16.4 μL P3 primary cell nucleofector supplement 3.6 μL Total 20 μL mTeSR Plus complete media
This should be freshly prepared and can be stored at 4 °C for two weeks.
Component Volume mTeSR Plus basal 40 mL mTeSR Plus 5× supplement 10 mL Total 50 mL Matrigel
Dilute Matrigel right before the experiment with DMEM/F12. The diluted Matrigel can be stored at 4 °C for 2 weeks.
Component Volume DMEM F12 11 mL Matrigel (1 mg/mL) 1 mL Total 12 mL
Laboratory supplies
6-well culture plate (Thermo Scientific, catalog number: 12556004)
24-well plate (Thermo Scientific, catalog number: 12556006)
12-well plate (Thermo Scientific, catalog number: 12556005)
1.7 mL microtube (Genesee Scientific, catalog number: 24-282C)
1,250 μL tips (GeneMate, catalog number: P-1237-1250)
200 μL tips (Neptune, catalog number: 89140-902)
20 μL tips (GeneMate, catalog number: P-1237-20)
10 μL tips (GeneMate, catalog number: P-1234-10)
15 mL Falcon tubes (Sarstedt, catalog number: 62.554.205)
16-well Nucleocuvette Strip (Lonza, catalog number: V4XP3032)
Equipment
Nucleofector (Lonza, model: 4D)
Centrifuge (Sorvall, model: ST16R)
Gel imager (Bio-Rad, model: ChemiDocTM MP Imaging System)
Gel electrophoresis machine (Labnet, model: Enduro GelXL)
NanoDrop (Thermo Scientific, model: Nanodrop One)
PCR thermocycler (Bio-Rad, model: T100TM thermal cycler)
Microscope (EVOS, Advance Microscopy group)
Incubator (SANYO CO2 incubator)
Biosafety cabinet (Nuaire, model: LabGard, Class II, Type A2, Biological Safety Cabinet)
Hemocytometer (Hausser Scientific Horsham, model: Bright-Line)
Software and datasets
Synthego.com (https://design.synthego.com/#/)
IDT (https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM)
ImageJ (https://ij.imjoy.io/)
Procedure
文章信息
稿件历史记录
提交日期: Aug 20, 2024
接收日期: Oct 14, 2024
在线发布日期: Nov 5, 2024
出版日期: Dec 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Singh, A., Babu, S., Phan, M. and Yuan, S. H. (2024). CRISPR/Cas9-Based Protocol for Precise Genome Editing in Induced Pluripotent Stem Cells. Bio-protocol 14(24): e5141. DOI: 10.21769/BioProtoc.5141.
- Singh, A., Smedley, G. D., Rose, J. G., Fredriksen, K., Zhang, Y., Li, L. and Yuan, S. H. (2024). A high efficiency precision genome editing method with CRISPR in iPSCs. Sci Rep. 14(1): 9933.
分类
干细胞 > 多能干细胞 > 基于细胞的分析方法
生物科学 > 生物技术 > CRISPR/Cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link