- EN - English
- CN - 中文
Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
利用免疫荧光法原位检测光合生物中TOR激酶活性
(§Technical contact: ana.lando@inbiotec.conicet.gov.ar; gnoel@inbiotec.conicet.gov.ar) 发布: 2024年12月20日第14卷第24期 DOI: 10.21769/BioProtoc.5140 浏览次数: 1806
评审: Shuhei OtaMalgorzata LichockaAnonymous reviewer(s)
Abstract
The target of rapamycin (TOR) is a central hub kinase that promotes growth and development in all eukaryote cells. TOR induces protein synthesis through the phosphorylation of the S6 kinase (S6K), which, in turn, phosphorylates ribosomal S6 protein (RPS6) increasing this anabolic process. Therefore, S6K and RPS6 phosphorylation are generally used as readouts of TOR activity. Protein phosphorylation levels are measured by a western blot (WB) technique using an antibody against one specific phosphosite in cell extracts. However, at the tissue/cell-specific level, there is a huge gap in plants due to the lack of alternative techniques for the evaluation of TOR activity as there are for other organisms such as mammals. Here, we describe an in vivo protocol to detect S6K phosphorylation in tissues/cells of model photosynthetic organisms such as Arabidopsis thaliana and Chlamydomonas reinhardtii. Our proposed method consists of the immunolocalization of a phosphorylated target of TOR kinase using a fluorescent secondary antibody by confocal microscopy. The protocol involves four main steps: tissue/cell fixation, permeabilization, and incubation with primary and secondary antibodies. It is an easy technique that allows handling different samples at the same time. In addition, different ultrastructural cell markers can also be used, such as for nucleus and cell wall detection, allowing a detailed analysis of cell morphology. To our knowledge, this is the first protocol to detect TOR activity in situ in photosynthetic organisms; we consider that it will pave the research on the TOR kinase, opening new possibilities to better understand its complex signaling.
Key features
• The protocol is an easy and non-destructive method to detect S6K phosphorylation at the cellular level for plants and algae.
• First method for in situ immunolocalization of target proteins of TOR kinase in photosynthetic organisms.
Keywords: Arabidopsis (拟南芥)Graphical overview
Background
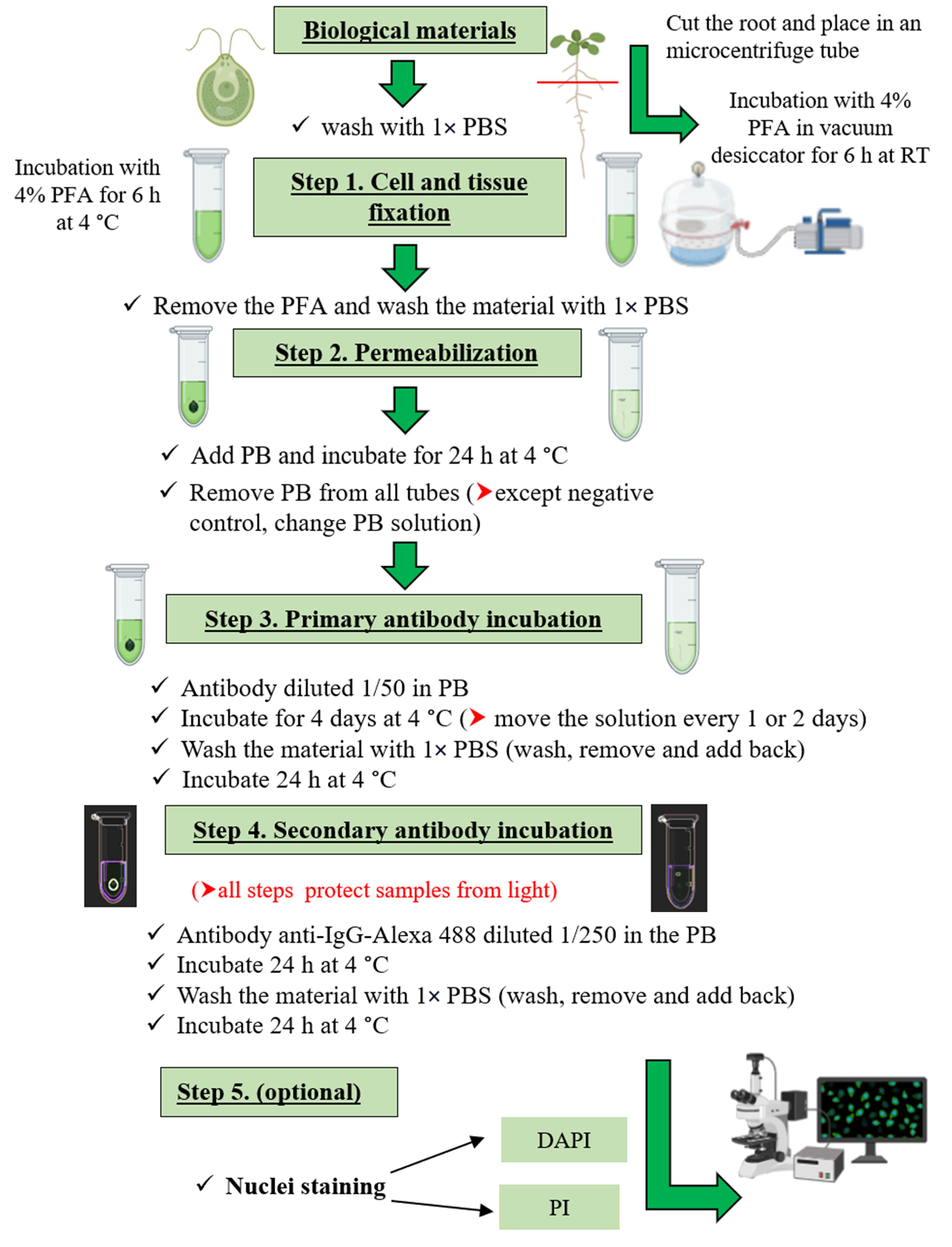
The target of rapamycin (TOR) kinase pathway is an ancestral signaling pathway that integrates nutrient information with translational control and growth regulation [1,2]. This conserved signaling pathway includes the S6K and its direct target, the 40S ribosomal protein S6 (RPS6) [3]. Therefore, the measurement of S6K and RPS6 phosphorylation levels through the WB technique is used as a reliable readout of TOR activity in eukaryotes including photosynthetic organisms [4,5]. The method is generally accurate in estimating TOR activity but requires cell disruption and does not permit subcellular localization or monitoring tissue specificities. Other techniques, such as immunofluorescence, have been described for mammals; however, in plants and algae, there is still a need to develop protocols for detecting TOR activity at the tissue/cell-specific level. Immunolocalization approaches can provide a great deal of information about the subcellular localization and dynamics of many plant proteins, even in vivo [6], which is particularly important when we have multi-level regulation, as is the case with the TOR kinase. We have developed, analyzed, and validated an immunofluorescence protocol for detecting TOR activity through S6K phosphorylation in situ in model photosynthetic organisms (Arabidopsis and Chlamydomonas). The main advantages of this method include its applicability to both plants and algae, its simplicity as it does not require tissue sectioning, its reproducibility due to Alexa's fluorophore stability, and the visualization in vivo due to the non-destruction of tissues/cells. Some minor limitations of the protocol include the restriction of its application to young tissues of plants, some reagents being expensive such as fluorescent antibodies, and the need for specialized personnel for the management of the confocal microscope.
This protocol will allow progress in the studies on TOR signaling at the subcellular level and tissue specificities, which are known to be essential for this pathway and where information is lacking in photosynthetic organisms, probably due to technical limitations.
To our knowledge, this is the first protocol for the analysis of TOR activity at the cellular level in plants and algae.
Materials and reagents
Biological materials
Arabidopsis thaliana Columbia ecotype (Col-0) wild-type (WT), young seedlings (5 days old)
Chlamydomonas reinhardtii CC125 (137c, mt + nit1 nit2), 15 mL of OD750nm: 0.6–0.9
Reagents
Double-distilled water (ddH2O)
Sodium phosphate dibasic (Na2HPO4) (Sigma, catalog number: 71496)
Potassium dihydrogen phosphate (KH2PO4) (Sigma, catalog number: 60220-M)
Sodium chloride (NaCl) (Anedra, catalog number: AN00716909)
Orthophosphoric acid (H3PO4) (Sigma, catalog number: 695017)
Paraformaldehyde (Agar Scientific, catalog number: R1018)
Sodium hydroxide (NaOH) (Fluka, catalog number: 71690)
Bovine serum albumin (BSA) 10 mg/mL (Promega, catalog number: R396D)
Triton X-100 (Sigma, catalog number: 9036-19-5)
Primary polyclonal antibody p-p70 S6K1 α Thr 389 (Santa Cruz, catalog number: sc-11759)
Secondary antibody anti-IgG-Alexa 488 (Invitrogen, catalog number: A11008)
Propidium iodide (PI) (Invitrogen, catalog number: P1304MP)
4',6-Diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma, catalog number: D9564)
Solutions
Paraformaldehyde (PFA) 4% (see Recipes)
Phosphate buffered saline (PBS) 5× (see Recipes)
Permeabilization and blocking buffer (PB) (see Recipes)
Recipes
PFA 4%
Weigh 4 g of PFA and place in 35 mL of double-distilled water at 60 °C, add 30–50 μL of 10 N NaOH, and leave to stir on a hot plate (until the PFA dissolves well).
Add 20 mL of 5× PBS.
Check pH; it should be between 7.2 and 7.8.
Bring up to 100 mL with ddH2O.
Split into Falcon tubes and store at -20 °C for up to 3 months and protected from light.
Caution: Highly toxic. Wear protective gloves and work in the fume hood when handling the powder.
PBS (5×) (for 1 L)
Dissolve 3.62 g of Na2HPO4 (FW 141.96), 1.05 g of KH2PO4 (FW 136.1), and 38.25 g of NaCl (FW 58.44) in 800 mL of ddH2O. Adjust the pH to 7.4 with H3PO4 and then add ddH2O to the final volume. Dispense the solution into aliquots and sterilize them by autoclaving. The work solution used is 1× (final concentration 8 mM Na2HPO4, 2 mM KH2PO4, and 140 mM NaCl).
Permeabilization and blocking buffer (PB) (for 1.5 mL of PB)
Add 150 μL of 1% BSA, 45 μL of 10% Triton X-100, and 1,305 μL of 1× PBS (final concentration 1× PBS, 0.1% BSA, and 0.3% Triton X-100).
Equipment
Confocal microscope (e.g., Nikon, model: Eclipse C1 Plus)
Vacuum pump with a desiccator
Software and datasets
Confocal microscope software (e.g., EZ-C1 software Nikon)
NIH ImageJ software 1.47 for Windows (https://imagej.net/ij/)
Procedure
文章信息
稿件历史记录
提交日期: Jun 19, 2024
接收日期: Oct 13, 2024
在线发布日期: Nov 5, 2024
出版日期: Dec 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Lando, A. P., De Marco, M. A., Cumino, A. C. and Martínez-Noël, G. M. A. (2024). Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms. Bio-protocol 14(24): e5140. DOI: 10.21769/BioProtoc.5140.
分类
植物科学 > 植物生物化学 > 蛋白质 > 活性
生物化学 > 蛋白质 > 免疫检测 > 免疫染色法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link