- EN - English
- CN - 中文
An Autocatalytic Platform Combining a Nonlinear Hybridization Chain Reaction and DNAzyme to Detect microRNA
结合非线性杂交链反应与DNA酶的自催化平台用于微小RNA检测
(*contributed equally to this work) 发布: 2024年12月20日第14卷第24期 DOI: 10.21769/BioProtoc.5134 浏览次数: 2037
评审: Han Deng Anonymous reviewer(s)

相关实验方案
新鲜冷冻骨骼肌切片上小鼠肌肉干细胞的RNA荧光原位杂交与免疫荧光染色同步检测
Vedant R. Lakkundi [...] Albert E. Almada
2025年09月05日 1912 阅读
Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that play pivotal roles in gene regulation; they are increasingly recognized as vital biomarkers for various diseases, notably cancer. Conventional methods for miRNA detection, such as quantitative PCR and microarray analysis, often entail intricate sample preparation and lack the requisite sensitivity to detect low-abundance miRNAs like miRNA-21. This protocol presents an innovative approach that combines branched hybridization chain reaction (bHCR) with DNAzyme technology for the precise detection of miRNA-21. The bHCR amplifies the target signal through a branched structure, while the DNAzyme boosts detection sensitivity through catalytic cleavage, enabling swift and specific identification of miRNA-21. This dual amplification strategy offers a highly sensitive, specific, and rapid alternative to traditional techniques, making it particularly well-suited for early-stage disease diagnosis.
Key features
• This protocol enables a sensitive detection of miRNA-21.
• The technique employs an isothermal, enzyme-free bHCR process that can be carried out using standard laboratory equipment, eliminating the necessity for specialized instruments.
• The entire protocol can be finalized in less than five hours, offering a swift and effective approach for high-throughput miRNA detection.
• Minimal input RNA is needed for the protocol, and the sample preparation steps are straightforward.
Keywords: DNAzyme (DNA酶)Graphical overview
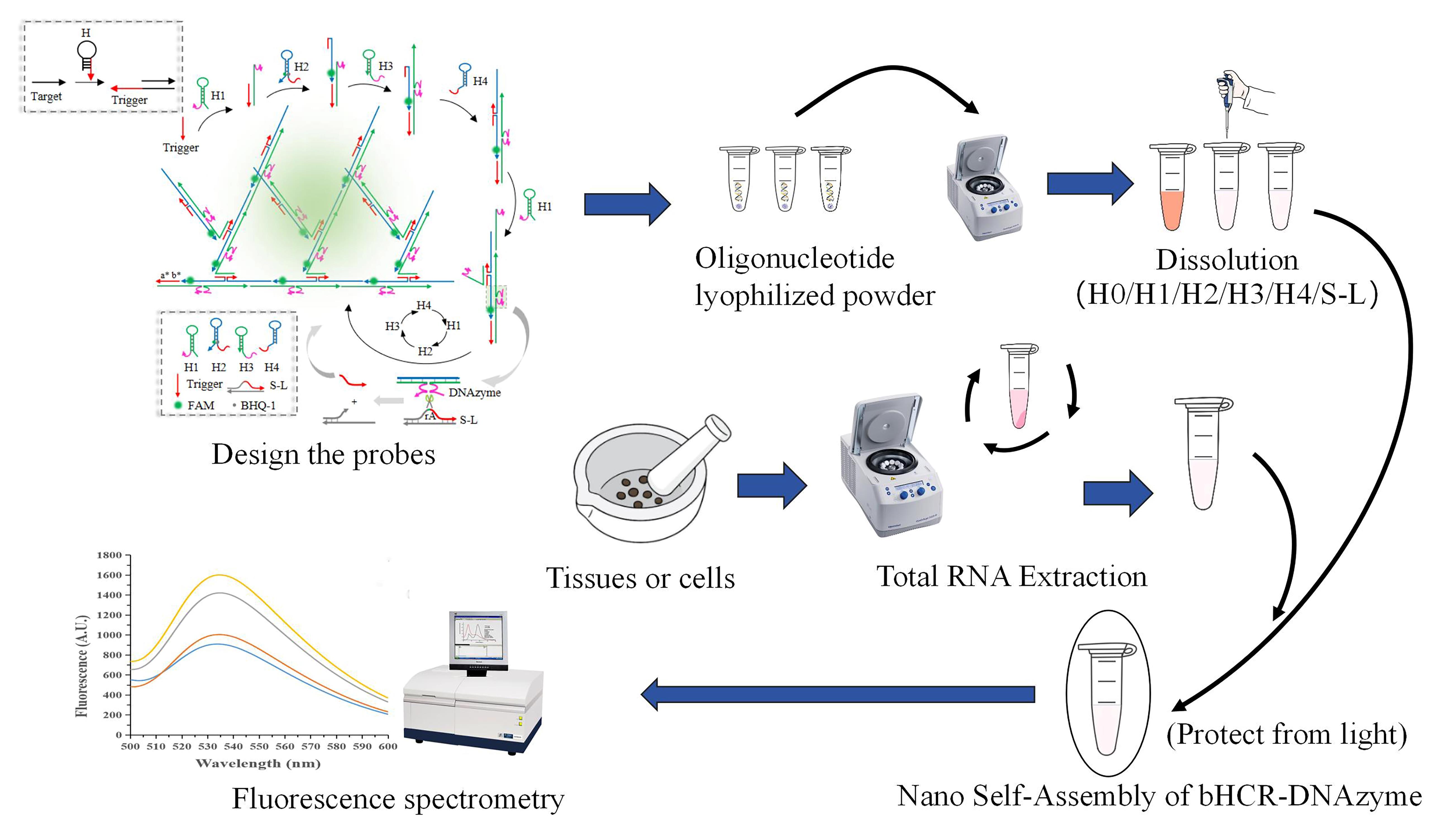
Detection of microRNA-21 based on bHCR-DNAzyme platform.
Note: For a more detailed explanation of the construction and design principles of this platform, please refer to the previously published article [1].
Background
MicroRNAs (miRNAs) are a class of small non-coding RNAs that play a crucial role in gene expression regulation by targeting messenger RNA (mRNA) for degradation or translational repression, thereby influencing various cellular processes [2]. In numerous diseases, miRNA expression patterns are dysregulated. For instance, miR-21 is upregulated in various cancers, including liver, breast, lung, and colorectal cancer. Conversely, the let-7 family of miRNAs, recognized for their tumor-suppressive roles, is often downregulated in numerous cancer types [3]. As a result, miRNAs are significant biomarkers closely associated with the development of various diseases, including cancer [4–7]. Detecting nucleic acid biomarkers is crucial for disease identification, treatment, and intervention. However, traditional miRNA detection methods face challenges due to the low abundance and high sequence homology of nucleic acid biomarkers, as well as limited detection sensitivity and complex, time-consuming procedures [8,9]. These limitations significantly impede the speed of research and clinical diagnostics. Hence, there is an urgent need to develop a novel method that combines sensitivity, specificity, and operational simplicity.
This protocol introduces a novel detection strategy that combines the synergistic effects of branched hybridization chain reaction (bHCR) and DNAzyme technology. In bHCR, fuel probes quickly assemble into networked or branched nanostructures upon the presence of an initiator, leading to exponential signal amplification. This feature enables the sensitive detection of low-abundance molecules and accelerates reaction speed [10]. However, bHCR has limitations that restrict its application, such as an upper limit to amplification, signal saturation, and increased background signal due to the spontaneous opening and self-amplification of hairpin probes [11]. On the other hand, DNAzyme is a unique nucleic acid catalytic enzyme with biological catalytic activity, such as RNA (or DNA) cleavage and ligation [12], obtained through in vitro selection [13]. It is known for its ease of synthesis, modifiability, and cost-effectiveness. With its separation of the recognition sequence and catalytic core, DNAzyme allows for flexible sequence design. Moreover, DNAzyme-catalyzed nucleic acid amplification mimics enzymatic reactions, theoretically enabling unlimited signal amplification [14,15]. However, DNAzyme exhibits a relatively slow linear amplification rate, and its enzymatic reactions can be influenced by product accumulation and enzyme inactivation [12]. By combining bHCR and DNAzyme technologies, this protocol harnesses the strengths of both methods to overcome their individual limitations, resulting in a novel nucleic acid detection approach for miRNA. Table 1 provides a comparative analysis of the advantages and disadvantages of both techniques. Among various nucleic acid biomarkers, miRNA-21 is notably upregulated in several cancers. Its elevated levels are strongly associated with tumor progression, metastasis, and poor patient prognosis [16–18], highlighting its potential as both a biomarker and a therapeutic target for early cancer detection. Therefore, this protocol uses miRNA-21 as an example to demonstrate how the bHCR-DNAzyme platform can detect miRNAs from tissues or cells. This dual amplification system offers a highly sensitive and specific method for detecting miRNA-21, even at low concentrations, in complex biological samples. The integration of bHCR and DNAzyme addresses various constraints of current miRNA detection techniques, potentially leading to more reliable and user-friendly early cancer diagnostics. The outcomes of this study could have significant clinical implications, enhancing the accessibility of clinical diagnostics and disease monitoring.
Table 1. Overview of bHCR and DNAzyme
| Branched hybridization chain reaction (bHCR) | Deoxyribozyme (DNAzyme) | |
|---|---|---|
| Concepts and principles | By triggering DNA hairpin structures, branched double-stranded DNA can be formed, enabling exponential amplification of nucleic acids. | A novel nucleic acid tool enzyme with biocatalytic activity, screened and isolated in vitro. |
| Advantages | • Isothermal amplification, requiring simple equipment. • Enzyme-free amplification, low cost, easy reagent storage, good reproducibility. • Exponential signal growth, rapid and strong signal output. | • Easy to synthesize, easy to modify, and cost-effective. • Separate recognition sequence and catalytic center, offering flexible design. • Comparable to enzymatic reactions, with no upper limit on signal amplification. |
| Disadvantages | • Amplification has an upper limit; signals can quickly saturate. • Prone to spontaneous amplification, leading to high background signals. | • Linear reaction, slower speed. • Product inhibition and enzyme inactivation require additional enzymes for long-term reactions. |
Note: The principles of the two techniques are illustrated in Figure 1.
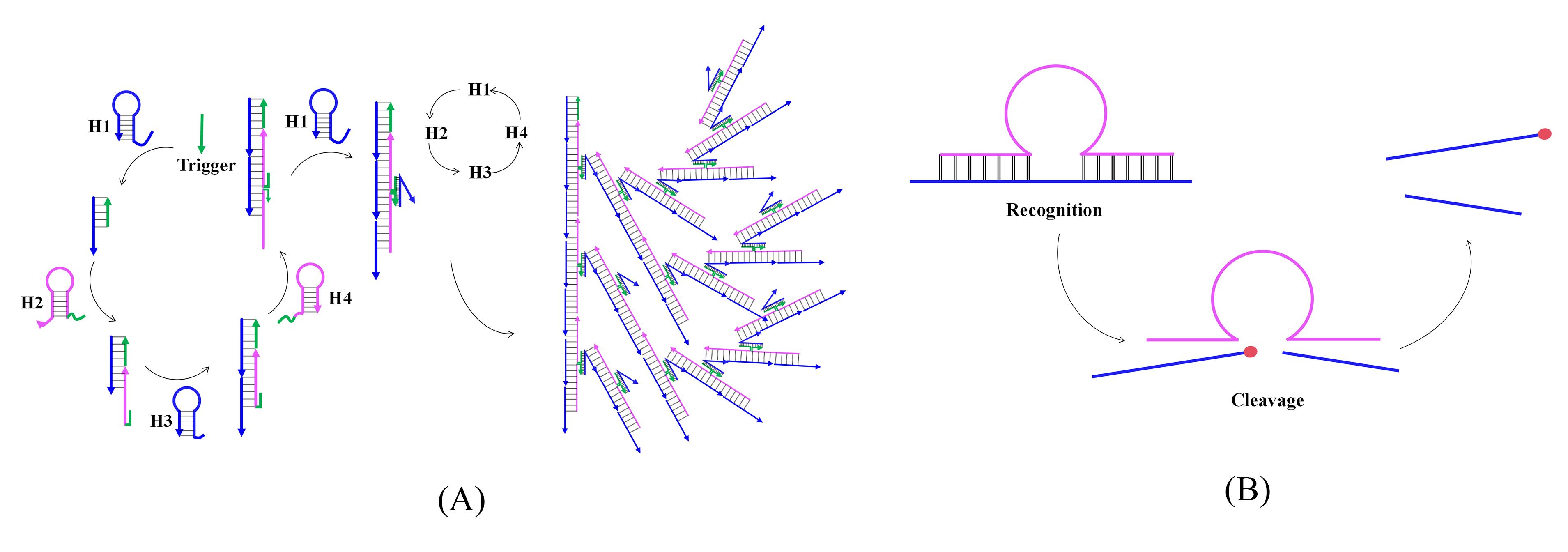
Figure 1. Principles of bHCR (A) and DNAzyme (B) techniques
Materials and reagents
Biological materials
Tumor and paratumor (PT) tissue from patients with hepatocellular carcinoma (from Third Xiangya Hospital in Central South University, Approval No: 2023009), stored in liquid nitrogen until analysis
Reagents
CaCl2 (Kermel Chemical Reagent Co Ltd., CAS: 10043-52-4)
NaCl (Kermel Chemical Reagent Co Ltd., CAS: 7647-14-5)
DEPC water (Biosharp, catalog number: BL510B)
HCl (Sinopharm, CAS: 7647-01-0)
1 M HEPES solution (Servicebio, catalog number: G4210-100ML)
Trizol (Jiangsu Cowin Biotech, catalog number: CW0580S)
Chloroform (Sinopharm, CAS: 67-66-3)
Isopropanol (Sinopharm, CAS: 67-63-0)
Ethanol (Sinopharm, Brand, CAS: 64-17-5)
MiRNA 1st strand cDNA synthesis kit (by tailing A) (Vazyme, Specification: 20rxns, catalog number: MR201-01)
Hieff® qPCR SYBR Green Master Mix (No Rox) (Yeasen, catalog number: 11201ES03)
10× TBE solution (Solarbio, catalog number: T1051)
30% Acr-Bis (29:1) (Coolaber, catalog number: SL1090)
10% APS (Coolaber, catalog number: SL1130)
TEMED (Coolaber, CAS: 110-18-9)
Labgreen (Coolaber, catalog number: SL2150)
6× loading buffer (Coolaber, catalog number: SL2210)
RNase-free water (Shanghai Promega, catalog number: SP119A)
Oligonucleotides (Beijing Tsingke Biotech, purification method: HPLC) (see Table 2 for details)
Table 2. Nucleic acid sequences used in this work
Note: The fluorescent group FAM and the quencher group BHQ-1 are both modified on the thymine (T) base; rA indicates that the DNAs were replaced by RNAs. The underlined bases in 1-mut, 2-mut, and 4-mut are mutant bases.Name Sequence (5′-3′) Trigger AGTCTAGGATTCAGCGTGGGATTA S CTCGGCACAAGTGGGTACATT rA GAGTCTAGGATTCAGCGTGGGATTA S1 CTCGGCACAAGTGGGTACATTAGAGTCTAGGATTCAGCGTGGGATTA L TAATCCGACCCTGATATACCACTTGTGCCGAG DNAzyme TAGACTTTCTCACAGCGTACTCGCTAAGGTTGTATGTAC H1 TAATCCCACGCTGAATCCTAGACTCAAAGTAGTCTAGGATTCAGCGT GCGCTAAGGTTGTATGTAC H2 AGTCTAGGATTCAGT(BHQ-1)CTAGGATTCAGCGTGCATCTCCACGCT GAATCCTAGACT(FAM)ACTTTG H3 TAGACTTTCTCACAGCGGAGATGCACGCTGAATCCTAGACTTCCA GGAGTCTAGGATTCAGCGTG H4 AGTCTAGGATTCAGCGTGGGATTACACGCTGAATCCTAGA CTCCTGGAAGCGTGGGATTA miR-21 UAGCUUAUCAGACUGAUGUUGA 1-mut UAGCUUAUCAAACUGAUGUUGA 2-mut UAACUUAUCAGACUAAUGUUGA 4-mut UAGAUUAUCAAACUGAUAUUAA H-miR-21 CTGAATCCTAGACTTCAACATCAGTCTGATAAGCTAAAAAGTCT AGGATTCAGCGTGGGATTA
Solutions
10 mM HEPES buffer (see Recipes)
75% ethanol solution (see Recipes)
1× TBE electrophoresis buffer (see Recipes)
2 µM SL initial concentration solution (see Recipes)
Recipes
10 mM HEPES buffer (100mL)
Reagent Final concentration Quantity CaCl2 100 mM 1.110 g NaCl 150 mM 0.877 g 1 M HEPES solution 10 mM n/a DEPC H2O n/a n/a Adjust the pH to 7.2 with HCl n/a n/a Total (optional) n/a 100 mL 75% ethanol solution (10 mL)
Reagent Final concentration Volume Anhydrous ethanol 75% (v/v) 7.5 mL H2O n/a see note Total n/a 10 mL Note: The water is made using an ultrapure water machine.
1× TBE electrophoresis buffer (500 mL)
Reagent Final concentration Volume 10× TBE n/a 50 mL H2O n/a see note Total n/a 500 mL Note: The water is made using an ultrapure water machine.
2 µM SL initial concentration solution (500 µL)
Reagent Final concentration Volume 100 µM S stock solutions 2 µM 2 µL 100 µM L stock solutions 2.5 µM 2.5 µL DEPC H2O n/a 95.5 µL Total n/a 100 µL Note: Prepare 100 µM stock solutions of S and L according to the provided oligonucleotide synthesis instructions. For sequence information, manufacturer and catalog number, please refer to item 19 in the Reagents section. The preparation process is outlined in step B2 of the Procedure.
Laboratory supplies
0.2 mL microcentrifuge tube (Biosharp, catalog number: BS-02-P)
1.5 mL microcentrifuge tube (Biosharp, catalog number: BS-15-M)
2 mL microcentrifuge tube (Biosharp, catalog number: BS-20-M)
15 mL centrifuge tube (Servicebio, catalog number: EP-5001-J)
50 mL centrifuge tube (Servicebio, catalog number: EP-5001-J)
10 μL pipette tip (Servicebio, catalog number: P-10)
200 μL pipette tip (Servicebio, catalog number: TP-200)
1,000 μL pipette tip (Servicebio, catalog number: TP-1250)
Pipette (China Dalong Xingchuang Experimental Instrument Co., Ltd, model: TopPette)
Cell freezing tube, 2 mL (Corning, catalog number: CLS430659)
Equipment
High-speed freezing centrifuge (Changsha Yingtai, model: TGL16)
Ultrapure water machine (Bertone, model: K.L.MINI4-T)
PCR amplifier (Bio-Rad, model: Biometra)
Ultramicro ultraviolet spectrophotometer (Qua Well, model: Q5000)
Vortex mixer (Shanghai Huxi, model: XW-80A)
Quartz fluorescent micro cuvette, 1 mm (Wuxi edge spectrum optics)
Biochemical incubator (Shanghai Boxun medical, model: SPX-150B-Z)
Electrophoresis analyzer (Beijing Liuyi Instrument Factory, model: DYY-7C)
Vertical electrophoresis tank (Beijing Junyi Oriental electrophoresis equipment Co., Ltd, model: DYCP-31BN)
Electronic balance (Shanghai Guangzheng medical, model: YP-B2002)
Fluorescence spectrophotometer (HITACHI, model: F-7100)
Real-time fluorescent quantitative gene amplification instrument (Bio-Rad, model: CFX CONNECT)
Clean bench (Suzhou Zhijing purification equipment Co., Ltd, model: SW-CJ-2FD)
Chemiluminescence imager (Bio-Rad, model: ChemiDoc XRS+)
Micro analytical balance (Shimadzu, model: AUW120D)
Software and datasets
GraphPad Prism (version 7, GraphPad Software)
Procedure
文章信息
稿件历史记录
提交日期: Aug 27, 2024
接收日期: Oct 10, 2024
在线发布日期: Nov 4, 2024
出版日期: Dec 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhang, H., Cao, X. and Zhu, Q. (2024). An Autocatalytic Platform Combining a Nonlinear Hybridization Chain Reaction and DNAzyme to Detect microRNA. Bio-protocol 14(24): e5134. DOI: 10.21769/BioProtoc.5134.
Cao, X., Dong, J., Sun, R., Zhang, X., Chen, C. and Zhu, Q. (2023). A DNAzyme-enhanced nonlinear hybridization chain reaction for sensitive detection of microRNA. J Biol Chem. 299(6): 104751.
分类
分子生物学 > RNA > RNA 检测
分子生物学 > RNA > qRT-PCR
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link