- EN - English
- CN - 中文
Quantitative Analysis of Kinetochore Protein Levels and Inter-Kinetochore Distances in Mammalian Cells During Mitosis
哺乳动物细胞有丝分裂中着丝点蛋白水平和着丝点间距的定量分析
(*contributed equally to this work) 发布: 2024年12月05日第14卷第23期 DOI: 10.21769/BioProtoc.5132 浏览次数: 1603
评审: Rajesh RanjanShanmugaPriyaa Madhukaran

相关实验方案

基于Fiji ImageJ的全自动化流程开发:批量分析共聚焦图像数据并量化蛋白共定位的Manders系数
Vikram Aditya [...] Wei Yue
2025年04月05日 2839 阅读
Abstract
The mammalian kinetochore is a multi-layered protein complex that forms on the centromeric chromatin. The kinetochore serves as the attachment hub for the plus ends of microtubules emanating from the centrosomes during mitosis. For karyokinesis, bipolar kinetochore-microtubule attachment and subsequent microtubule depolymerization lead to the development of inter-kinetochore tension between the sister chromatids. These events are instrumental in initiating a signaling cascade culminating in the segregation of the sister chromatids equally between the new daughter cells. Of the hundreds of conserved proteins that constitute the mammalian kinetochore, many that reside in the outermost layer are loaded during early mitosis and removed around metaphase-anaphase. Dynamically localized kinetochore proteins include those required for kinetochore-microtubule attachment, spindle assembly checkpoint proteins, various kinases, and molecular motors. The abundance of these kinetochore-localized proteins varies at prometaphase, metaphase, and anaphase, and is thus considered diagnostic of the fidelity of progression through these stages of mitosis. Here, we document detailed, state-of-the-art methodologies based on high-resolution fluorescence confocal microscopy followed by quantification of the levels of kinetochore-localized proteins during mitosis. We also document methods to accurately measure distances between sister kinetochores in mammalian cells, a surrogate readout for inter-kinetochore tension, which is essential for chromosome segregation.
Key features
• Immunostaining of cultured and suitably fixed adherent mammalian cells growing as monolayers.
• Confocal fluorescence imaging for the purpose of fluorescence quantification.
• 2D and 3D image reconstruction and analysis of the acquired images using appropriate background correction and normalization.
• Quantification of the inter-sister kinetochore distances using 3D image reconstruction.
Keywords: Mitosis (有丝分裂)Graphical overview
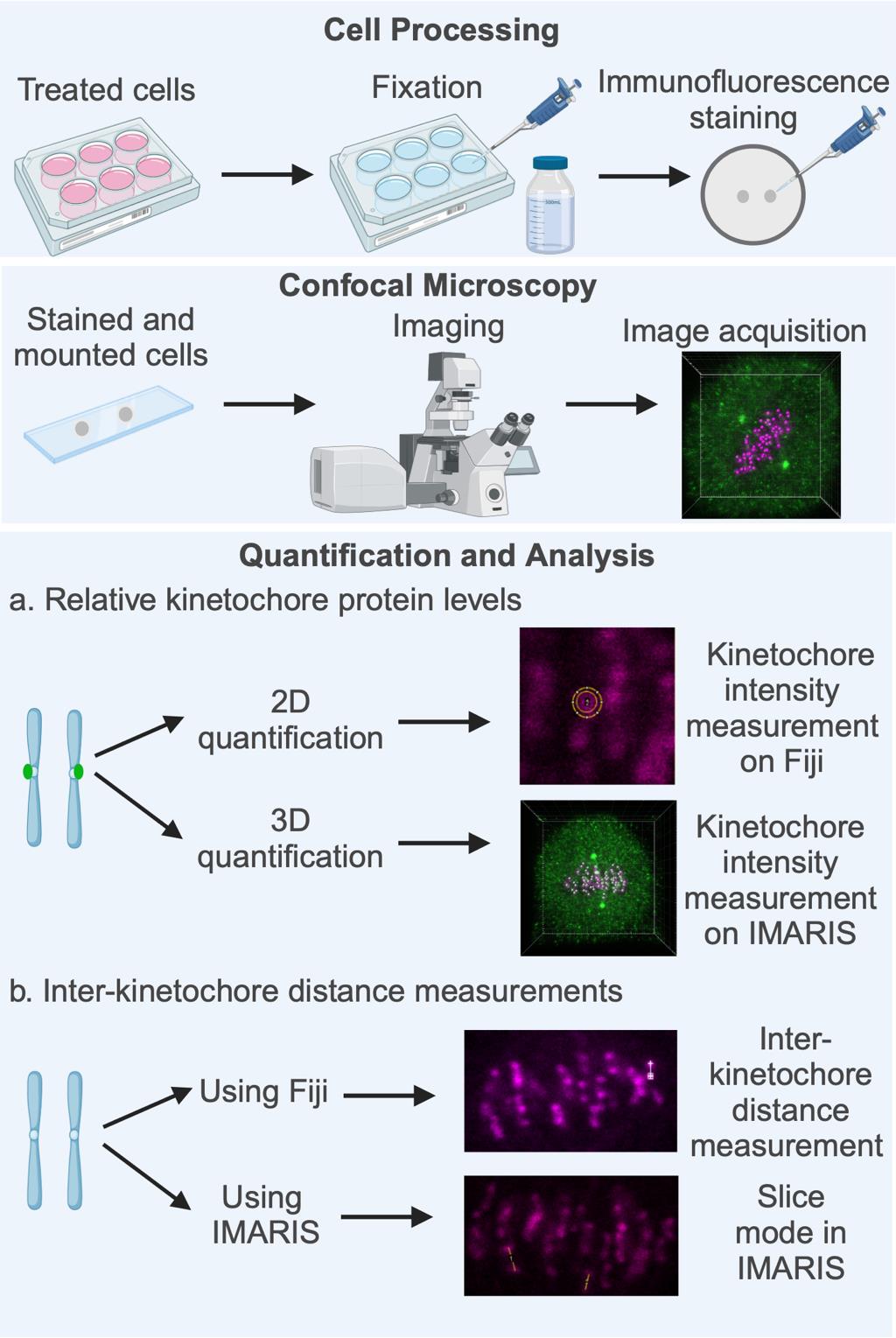
Key steps involved in fluorescence quantification of spindle assembly checkpoint (SAC) protein loading at the kinetochores and in the calculation of inter-sister kinetochore distances. High-resolution confocal image datasets of cells subjected to appropriate treatment as per the experimental need (drugs, inhibitors, gene-specific siRNA, etc.) are used for quantifying the levels of SAC proteins at the kinetochores, measuring inter-sister kinetochore distances, or both. Quantification and analysis of both parameters are performed using image analysis software such as Fiji (open source) or the IMARIS software suite. Figure created with BioRender.com.
Background
The spindle assembly checkpoint (SAC) is the primary mechanism that ensures equal sister chromatid segregation [1–3]. The SAC monitors two major parameters during mitosis. The first is the end-on attachment of centrosome-derived microtubule plus ends to the kinetochores (Kt-MT attachment). The establishment of bipolar (amphitelic) Kt-MT attachment of a sister chromatid pair to opposite centrosomes, followed by the depolymerization of these kinetochore microtubules (kMTs), leads to poleward pulling of the kinetochores and the generation of inter-kinetochore tension, the second parameter sensed by the SAC. All other stochastically occurring forms of Kt-MT mis-attachment (monotelic, merotelic, and syntelic) fail to elicit inter-kinetochore tension [1].
The SAC operates through SAC proteins resident on the fibrous corona, the outermost and most transient kinetochore layer. The SAC generates a diffusible “wait-anaphase” signal triggered by the lack of attachment or mis-attachment at even one kinetochore, stalling the entire cell in metaphase and providing time for the rectification of Kt-MT mis-attachments [3]. Inter-kinetochore tension stabilizes Kt-MT end-on attachments and also initiates a molecular signaling cascade instrumental for the separation of sister chromatids toward the opposite spindle poles (centrosomes) due to kMT depolymerization. Inactivation or silencing of the SAC, achieved primarily through dynein-based poleward “stripping” of SAC proteins from kinetochores along spindle microtubules upon Kt-MT attachment or tension, is essential for anaphase onset [1,3].
The level of enrichment of SAC proteins, their receptors, and other associated proteins at the kinetochores can serve as biochemical markers of the exact stage of mitosis or be used as indicators of specific defects in mitotic progression [4]. High-resolution confocal microscopy has been used to reliably quantify these levels and correlate them with the stage of mitosis, mainly because the size of human kinetochores is not optically diffraction-limited [5]. Kinetochore protein level quantification methods were initially developed on individual, two-dimensional confocal micrographs of single z-planes, taking into account local background fluorescence corrections around the kinetochores [5] and have been used extensively since. In recent years, this concept has been built upon to evolve widely used methods employing modern microscopy analysis software packages and three-dimensional image reconstructions [6–8]. Similar tools have been used to quantify inter-kinetochore distances at prometaphase and metaphase. These distance measurements have proven to be reliable correlates of inter-kinetochore tension, especially at late metaphase prior to segregation [1]. This article details state-of-the-art methodologies for the quantification of levels of kinetochore proteins and distance measurements between sister kinetochores. The methods described can be adapted to most cultured cells/cell lines that can be imaged using high-resolution confocal fluorescence microscopy.
Materials and reagents
Reagents
Immunofluorescence labeling
Primary antibodies against the protein(s) of interest (the Mad1 example is shown here):
Mad1 (Thermo Fisher Scientific, catalog number: PA5–28185). Other alternatives compatible with immunofluorescence staining may be used after empirical optimization.
CREST (to stain kinetochores) (Antibodies Incorporated, catalog number: 15–234). Other alternatives staining the kinetochore or the centromere that are compatible with immunofluorescence staining may be used after empirical optimization for specificity.
α-Tubulin, DM1 alpha (Sigma, catalog number: T9026)
Secondary antibodies tagged with fluorophore:
Alexa Fluor® 488 AffiniPure Donkey Anti-Rabbit IgG (Jackson ImmunoResearch Inc., catalog number: 711–545-152)
Alexa Fluor® 594 AffiniPure donkey Anti-Mouse IgG (Jackson ImmunoResearch Inc., catalog number: 715–585-150)
CyTM 5 AffiniPure Donkey Anti-Human IgG (Jackson ImmunoResearch Inc., catalog number: 709–175-149)
Note: Other alternative fluorophore-conjugated secondary antibodies compatible with immunofluorescence staining may be used after empirical optimization.
Sodium chloride (NaCl) (SRL, catalog number: 64072); may be procured commercially from other standard sources
Potassium chloride (KCl) (SRL, catalog number: 38630); may be procured commercially from other standard sources
Disodium hydrogen phosphate (Na2HPO4) (SRL, catalog number: 1949144); may be procured commercially from other standard sources
Potassium dihydrogen phosphate (KH2PO4) (SRL, catalog number: 1649201); may be procured commercially from other standard sources
Triton X-100 (Sigma, catalog number: T8787–100ML)
Bovine serum albumin (BSA) (pH 6–7) (SRL, catalog number 9048–46-8); may be procured commercially from other standard sources
4',6-diamidino-2-phenylindole (DAPI), 5 mg/mL stock solution made in dimethyl sulfoxide (DMSO) (Thermo Scientific, catalog number: 62247)
Note: Alternatively, chromatin dyes for immunofluorescence such as Hoechst 33342 may be used after empirical optimization.
ProLong DiamondTM mounting media (Thermo Scientific, catalog number: P36970). Alternative antifade mounting media with a refractive index compatible with the lens and the lens immersion medium may be used after empirical optimization
1× phosphate buffered saline (PBS) (see Recipes)
Blocking solution (see Recipes)
1× phosphate buffered saline (PBS)
Components Amount/volume added Working concentration NaCl 8 g 137 mM KCl 0.2 g 2.7 mM Na2HPO4 1.44 g 10 mM KH2PO4 0.24 g 2 mM Deionized water Make up the volume to 1 L Blocking solution for immunofluorescence staining
Components Amount/volume added Working concentration PBS 50 mL 1× Triton X-100 250 µL 0.5% BSA 0.5 g 1% Weigh 0.5 g of BSA and add to 45 mL of 1× PBS in a 50 mL tube.
Place the tube on a rocker and mix at slow speed until completely dissolved. Avoid frothing of the solution.
Caution: High-speed mixing can denature proteins including BSA, of which frothing is a sign.
Add 250 μL of Triton X-100 to the solution.
Slowly mix the solution on a rocker to dissolve all the components completely.
Add 1× PBS to make up the volume to 50 mL and gently mix to homogeneity.
Clean glass slides and coverslips
Critical: The coverslips should be properly cleaned and ideally be of category #1.5 (even thickness between 0.16 and 0.19 mm) to minimize chromatic aberration and refractive index mismatches between the lens, immersion medium, and sample mounting medium.
Equipment
Confocal image acquisition
Leica TCS SP8 laser scanning optical confocal microscope with HCX PL APO 63X-1.4 NA oil-immersion objective and a HyD (hybrid) or photomultiplier tube (PMT) detector.
Note: Equivalent confocal microscopes from standard commercial sources may be used, as long as they offer the following key features:
Plan apochromat (PL APO) confocal grade objectives with a high numerical aperture (1.2 or above).
Laser illumination in at least three distinct colors.
Emission pinhole(s) designed to block out-of-focus emission light from adjacent z-planes from being captured in the images.
In the case of raster laser scanning confocal scan-heads, high-sensitivity detectors such as PMTs, hybrid detectors, or avalanche photodiodes (APDs) with a quantum efficiency in the visible spectrum around 50 % may be used. Alternatively, for spinning disc confocal microscopes, high sensitivity cameras such as scientific complementary metal oxide semiconductor (sCMOS) cameras or electron multiplying charge-coupled device (EMCCD) cameras, with a quantum efficiency in the visible spectrum of around 80% or above, may be used.
Software and datasets
For image analysis
IMARIS software suite, version 8.3, Bitplane. Higher versions may be used.
Fiji (open-source image analysis software) (https://fiji.sc, Schindelin et al. [9])
For statistical analysis
GraphPad Prism (GraphPad Software, USA). Other standard software packages may be used.
Procedure
文章信息
稿件历史记录
提交日期: Apr 8, 2024
接收日期: Sep 30, 2024
在线发布日期: Nov 4, 2024
出版日期: Dec 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Wasnik, N., Singhal, M., Khantwal, S., Mylavarapu, S. and Mylavarapu, S. V. S. (2024). Quantitative Analysis of Kinetochore Protein Levels and Inter-Kinetochore Distances in Mammalian Cells During Mitosis. Bio-protocol 14(23): e5132. DOI: 10.21769/BioProtoc.5132.
- Kumari, A., Kumar, C., Pergu, R., Kumar, M., Mahale, S. P., Wasnik, N. and Mylavarapu, S. V. (2021). Phosphorylation and Pin1 binding to the LIC1 subunit selectively regulate mitotic dynein functions. J Cell Biol. 220(12): e202005184. https://doi.org/10.1083/jcb.202005184
分类
细胞生物学 > 细胞成像 > 共聚焦显微镜
细胞生物学 > 基于细胞的分析方法
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link