- EN - English
- CN - 中文
Compartment-Resolved Proteomics with Deep Extracellular Matrix Coverage
深度覆盖细胞外基质的分区蛋白质组学研究
发布: 2024年12月05日第14卷第23期 DOI: 10.21769/BioProtoc.5123 浏览次数: 1901
评审: Rama Reddy GoluguriNeha NandwaniAnonymous reviewer(s)

相关实验方案

适用于 LC–MS/MS 蛋白质组学分析的大体积细胞培养上清分泌组样品制备方法优化
Basil Baby Mattamana [...] Peter Allen Faull
2025年12月20日 1209 阅读
Abstract
The extracellular matrix (ECM) is a complex network of proteins that provides structural support and biochemical cues to cells within tissues. Characterizing ECM composition is critical for understanding this tissue component’s roles in development, homeostasis, and disease processes. This protocol describes an integrated pipeline for profiling both cellular and ECM proteins across varied tissue types using mass spectrometry–based proteomics. The workflow covers stepwise extraction of cellular and extracellular proteins, enzymatic digestion into peptides, peptide cleanup, mass spectrometry analysis, and bioinformatic data processing. The key advantages include unbiased coverage of cellular, ECM-associated, and core-ECM proteins, including the fraction of ECM that cannot be solubilized using strong chaotropic agents such as urea or guanidine hydrochloride. Additionally, the method has been optimized for reproducible ECM enrichment and quantification across diverse tissue samples. This protocol enables systematic mapping of the ECM at a proteome-wide scale.
Key features
• Improved profiling of core extracellular matrix and matrisome-associated proteins through multi-step decellularization and chemical extraction of insoluble ECM
• Extraction buffers optimized for effectiveness across a broad range of tissue types and compatibility with varied MS platforms
• Measurement of protein solubility via resistance to detergent and chaotrope extraction
• Integrated LC-MS/MS analysis and data processing pipeline for ECM-focused analysis
Keywords: proteomics (蛋白质组学)Graphical overview
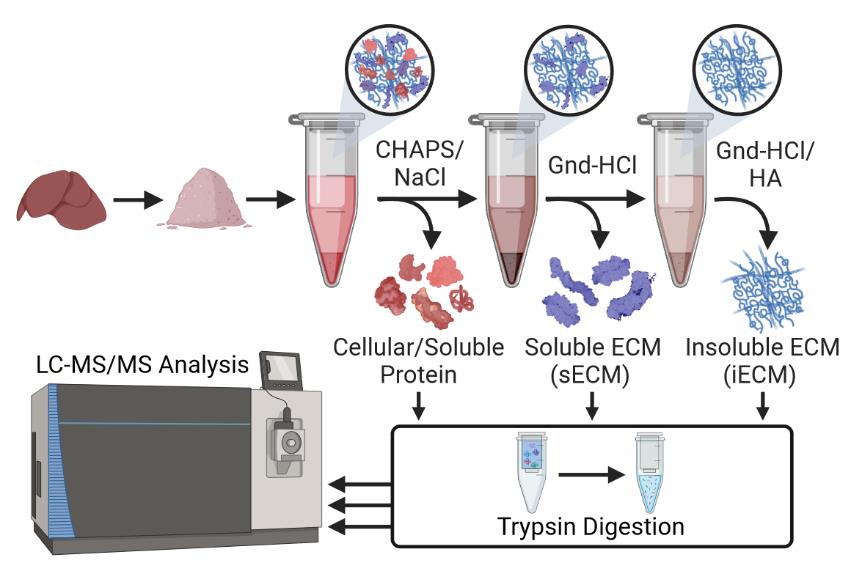
ECM-focused protein extraction and analysis workflow
Background
The extracellular matrix (ECM) is a complex biomolecular network composed of collagens, glycoproteins, proteoglycans, and other components that provide structural support and regulate the function of the ECM [1]. The ECM provides essential physical scaffolding for cellular constituents, as well as critical biochemical and biomechanical cues that regulate tissue development, homeostasis, and regeneration [2]. Given its fundamental roles, dysregulation of the ECM is implicated in numerous pathologies including fibrosis [3,4], cardiovascular diseases [5,6], and cancer progression [7]. Comprehensive mapping of ECM composition is crucial for understanding the diverse functions of this intricate network across different tissue microenvironments and disease states. Early efforts to catalog the ECM focused on profiling a subset of known ECM proteins [8] or employed antibody-based approaches [9], which are inherently biased and limited in coverage. More recently, mass spectrometry (MS)-based proteomics has enabled unbiased and system-wide profiling of the ECM at a proteome scale. However, technical challenges in extracting and enriching the ECM while minimizing contamination from cellular proteins have impeded comprehensive ECM characterization across a wide range of tissue types and organs. Previous analyses have shown that more than 80% of the total fibrillar collagen in many tissues resides in a chaotrope-resistant insoluble ECM fraction and is not solubilized by traditional proteomic extraction methods [10], highlighting the need for additional extraction steps when comprehensive characterization of ECM composition is a priority (Figure 1).
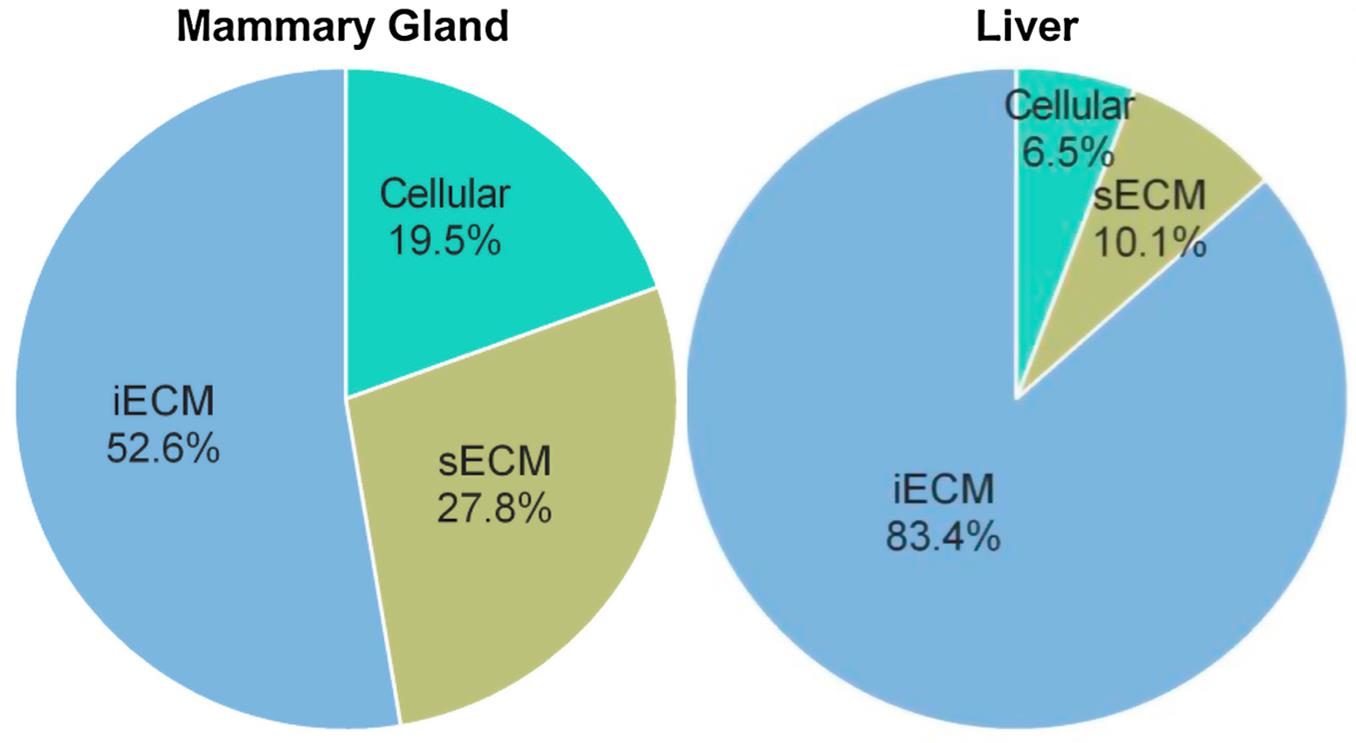
Figure 1. Proportion of total collagen I identified in the cellular, soluble extracellular matrix (sECM) and insoluble ECM (iECM, requiring chemical digestion) fractions of a 3-step ECM extraction via QconCAT-based absolute quantification. Adapted from Goddard et al. [10]. This protocol outlines an integrated experimental and computational pipeline tailored for deep proteomic profiling of the ECM across multiple mouse tissues and organs. The methods here have been refined across a broad range of tissue types, including 25 different mouse organs [11], and have been comprehensively benchmarked against other prominent methods in the field to validate their performance [12]. The workflow covers 1) enhanced step-wise extraction of cellular and ECM proteins facilitated by optimized extraction buffers effective across varied tissue types; 2) enzymatic digestion of proteins into peptides; 3) subsequent sample quantification and cleanup for MS analysis; 4) tandem mass spectrometry (LC-MS/MS) analysis of cellular and ECM protein fractions; and 5) bioinformatic identification of peptide sequences and computational matrisome categorization to classify ECM and associated proteins.
Materials and reagents
Reagents
Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) (Roche, catalog number: 10812846001)
3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS) (Sigma-Aldrich, catalog number: C3023)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E4884)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888)
Halt protease inhibitor cocktail (Thermo Fisher, catalog number: 78429)
Guanidine hydrochloride (Gnd-HCl) (Sigma-Aldrich, catalog number: G4505)
Hydroxylamine-HCl (NH2OH–HCl) (Sigma-Aldrich, catalog number: 159417)
Ammonium bicarbonate (ABC) (Sigma-Aldrich, catalog number: A6141)
Double-distilled water (ddH2O)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9779)
Iodoacetamide (IAM) (Sigma-Aldrich, catalog number: I1149)
Potassium carbonate (K2CO3) (Sigma-Aldrich, catalog number: 209619)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S5881)
Urea (Sigma-Aldrich, catalog number: U5128)
Buffer A: 0.1% formic acid (FA) in H2O, Optima (Thermo Fisher, catalog number: LS118)
Buffer B: 0.1% FA in 80% ACN, Optima (Thermo Fisher, catalog number: LS122500)
Sequencing-grade modified trypsin (Promega, catalog number: V5111)
Lysyl Endopeptidase®, mass spectrometry grade (Lys-C) (FUJIFILM Wako Pure Chemical Corporation, catalog number: 125-05061)
Optional (if necessary based on protocol notes):
Gel-loading pipette tips (Corning, catalog number: CLS4853)
OptimaTM acetone (Thermo Fisher, catalog number: A929)
PierceTM 660 nm protein quantification assay (Thermo Fisher, catalog number: 22660)
High salt buffer (see Recipes)
Gnd-HCl solution (see Recipes)
DTT solution (see Recipes)
IAM solution (see Recipes)
Hydroxylamine (HA) buffer preparation (pH 9.0) (see Recipes)
Urea solution (see Recipes)
Digest master mix (per sample) (see Recipes)
High salt buffer
Reagent Final concentration Amount Tris-HCl (1M, pH 7.4) 50 mM 5 mL CHAPS 0.25% 25 mg EDTA (500 mM) 25 mM 5 mL NaCl 3 M 17.53 g Halt protease inhibitor (100×) 1× 1 mL Total n/a 100 mL Combine buffer components and bring to 100 mL with ddH2O. Buffer without protease inhibitors can be prepared in advance and stored at 4 °C for up to one month. Protease inhibitors must be added fresh before use.
Gnd-HCl solution
Reagent Final concentration Amount Gnd-HCl 6 M 57.32 g ABC (1 M, pH 8.0) 100 mM 10 mL Total n/a 100 mL Dissolve Gnd-HCl in ddH2O before adding 10 mL of 1 M ABC. Adjust pH to 9.0 using NaOH. Add ddH2O to 100 mL. Prepare fresh before use.
DTT solution
Reagent Final concentration Amount DTT 100 mM 15.4 mg ABC (1 M) 100 mM 1 mL Total n/a 10 mL Combine components and add ddH2O to 10 mL. Prepare fresh before use.
IAM solution
Reagent Final concentration Amount IAM 500 mM 92.5 mg ABC (1 M) 100 mM 1 mL Total n/a 10 mL Combine components and add ddH2O to 10 mL. Prepare fresh before use and store in the dark.
Hydroxylamine (HA) buffer preparation (pH 9.0)
Reagent Final concentration Amount Gnd-HCl solution (Recipe 2) 70% v/v 70 mL NH2OH–HCl 1 M 7 g NaOH (50% w/v) 4% w/v 8 mL K2CO3 (1 M) 0.2 M 20 mL Total n/a 100 mL Prepare Gnd-HCl solution according to Recipe 2. Dissolve NH2OH–HCl in 50 mL of Gnd-HCl solution. Add NaOH, mix, then add K2CO3. Add Gnd-HCl solution to 90 mL before adjusting pH to 9.0 using HCl. Add Gnd-HCl solution to 100 mL. Prepare fresh before use.
Urea solution
Reagent Final concentration Amount Urea 8 M 24.02 g ABC (1 M) 100 mM 5 mL Total n/a 50 mL Combine components and add ddH2O to 50 mL. Prepare fresh before use.
Digest master mix (per sample)
Reagent Final Concentration Amount ABC (1 M) 25 mM 2.5 μL Lys-C 1:400 enzyme:protein ratio 0.075 μg (for 30 μg protein) Total n/a 100 μL
Laboratory supplies
MS-compatible pipette tips (any brand)
Safe-Lock 1.75 mL tubes (Eppendorf, catalog number: 003012361)
2 mL snap cap tubes (any brand)
Glass beads, 1 mm diameter (Next Advance, catalog number: GB10)
10 kDa molecular weight cutoff (MWCO) filters (Sartorius, catalog number: VN01H02)
PierceTM quantitative colorimetric peptide assay (Thermo Fisher, catalog number: 23275)
Flat-bottom, clear 96-well plate (Grenier, catalog number: 655101)
Empore sealing tape for 96-well plate (Millipore Sigma, catalog number: 66881-U)
PierceTM C18 spin tips (Thermo Fisher, catalog number: 84850)
Verex 300 μL vials with pre-slit cap (Phenomenex, catalog number: AR0-39S0-13-C/AR0-8972-13-C)
Equipment
Ceramic mortar and pestle (any brand)
Measuring pipettes, p1000, p200, p10 (any brand)
FreeZone 4.5L benchtop freeze dryer (Labconco, catalog number: 7750020)
Clear complete fast-freeze flask, any size (Labconco)
Scroll vacuum pump (Labconco, catalog number: 7587000)
Bullet blender (Next Advance, model: BBX24)
Centrifuge (Eppendorf, model: 5430R)
Vortex mixer with 1.5 mL tube attachment (Labnet, model: VX-200)
Orbital shaker, Orbit M60 (Labnet, catalog number: LS-2020-M60)
Spark® microplate reader (TECAN, catalog number: 30086376)
CentriVap vacuum concentrator (Labconco, catalog number: 7810016)
Fusion Lumos Tribrid mass spectrometer (Thermo Fisher)
EASY-nLC 1200 System (Thermo Fisher, catalog number: LC140)
Analytical column made in-house: 100 μm i.d. × 150 mm fused silica capillary packed with 2.7 μm CORTECS C18 resin (Waters). Commercial alternative: EASY-Spray C18 column, 75 μm i.d. × 150 mm fused silica capillary packed with 3 μm resin (Thermo Fisher, catalog number: ES900)
Software and datasets
XCalibur Version 4.5 (Thermo Fisher)
Mascot Server Version 2.5 (Matrix Science)
Proteome Discoverer Version 2.5 (Thermo Fisher)
MSFragger (Nesvizhskii Lab) [13]
UniProtKB (https://www.uniprot.org/help/uniprotkb)
CRAPome Contaminant Database [14]
MatrisomeDB [15]
Procedure
文章信息
稿件历史记录
提交日期: May 1, 2024
接收日期: Sep 23, 2024
在线发布日期: Oct 22, 2024
出版日期: Dec 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
McCabe, M. C., Saviola, A. J. and Hansen, K. C. (2024). Compartment-Resolved Proteomics with Deep Extracellular Matrix Coverage. Bio-protocol 14(23): e5123. DOI: 10.21769/BioProtoc.5123.
分类
系统生物学 > 蛋白质组学 > 分泌蛋白质组
生物化学 > 蛋白质 > 定量
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link