- EN - English
- CN - 中文
A High-Throughput Droplet-based Method to Facilitate Microbial Conjugation
基于微滴的高通量方法促进微生物接合
(*contributed equally to this work, § Technical contact) 发布: 2024年12月05日第14卷第23期 DOI: 10.21769/BioProtoc.5120 浏览次数: 1615
评审: Alba BlesaMercedes SanchezFernando A Gonzales-Zubiate

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1231 阅读
Abstract
Droplet microfluidic platforms have been broadly used to facilitate DNA transfer in mammalian and bacterial hosts via methods such as transformation, transfection, and conjugation, as introduced in our previous work. Herein, we recapitulate our method for conjugal DNA transfer between Bacillus subtilis strains in a droplet for increased conjugation efficiency and throughput of an otherwise laborious protocol. By co-incubating the donor and recipient strains in droplets, our method confines cells into close proximity allowing for increased cell-to-cell interactions. This methodology is advantageous in its potential to automate and accelerate the genetic modification of undomesticated organisms that may be difficult to cultivate. This device is also designed for modularity and can be integrated into a variety of experimental workflows in which fine-tuning of donor-to-recipient cell ratios, growth rates, and media substrate concentrations may be necessary.
Key features
• Builds on previous Bacillus-conjugation methods introduced by Brophy et al. [1] increasing the throughput by flowing the donors and recipients into a droplet microfluidic chip.
• Experiments performed on this chip increase conjugation efficiency as compared to conjugation performed traditionally by co-incubating the cells in free culture.
• This platform enables fine-tuning of experimental parameters, e.g., donor-to-recipient cell ratios, induction concentration, and incubation times, all critical factors in engineering undomesticated organisms.
• Adaptable for upstream automation of bacterial cultivation and downstream analysis of transconjugants encapsulated in droplets.
Keywords: Droplet microfluidics (微滴微流控)Graphical overview
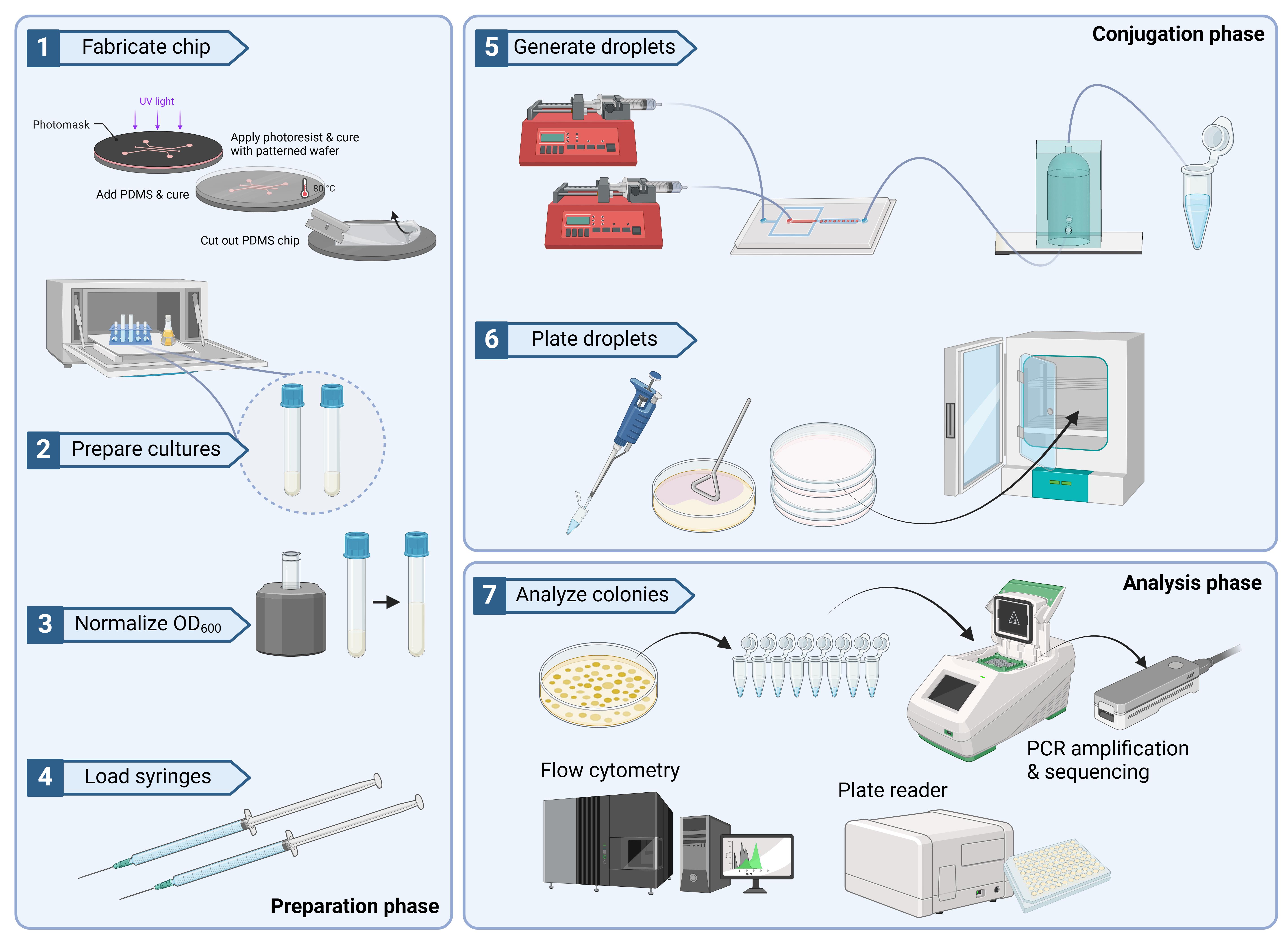
Background
Droplet microfluidic platforms have been broadly used to enable facile transformation of both mammalian and microbial cells in a high-throughput manner. This has been demonstrated through microfluidic electroporation of droplet-encapsulated cells [2,3], chemically-mediated transformation of cells (in some instances with heat shock added) [4,5], as well as transfection through means of mechanoporation [6], which has even been demonstrated in difficult-to-transfect primary T lymphocytes [7], moving toward CAR-T applications.
As evidenced, there exists a wide array of droplet-based microfluidic tools and techniques developed to streamline transformation and transfection, yet conjugative methods of DNA transfer have been scarcely explored. While conjugation is a modality of DNA transfer exclusive to microbial species, it is a powerful method commonly used to introduce heterologous DNA into microbial hosts that may be less amenable to modification. A canonical example of this is E. coli-based conjugation between F+ and F- cells. In this case, E. coli containing an F plasmid are able to synthesize a pilus structure allowing for DNA to be transferred during cell-to-cell contact [8]. Conversely, Agrobacterium-mediated DNA transfer, which leverages its native pathogenic functions to transfer DNA to a microbial or plant host, is able to do so through a Type IV secretion system [9]. Lastly, a unique example of conjugation is that of some strains of B. subtilis, which involves the transfer of integrative conjugative elements (ICE) from one host to the other. The XPORT strain [1], used in this protocol, is an engineered donor strain with a modified ICE. In the aforementioned work, this strain was used to transfer DNA via conjugation to a broad array of undomesticated strains of Bacillus and other related strains. Moreover, XPORT has demonstrated survivability and successful conjugation in soil environments where cell growth is uncontrolled. While the biological components of the system are robust, we have designed a microfluidic platform (previously introduced in Wippold et al. [10]) that offers more experimental control and complements the workflow described by Brophy et al. [1], expediting an otherwise laborious protocol and enabling easy optimization of experimental parameters such as donor-to-recipient ratios.
Although there have been other efforts demonstrating in-droplet conjugation, such as that introduced by Lam et al. [11], we propose two advantages to our protocol: (1) The lower surfactant concentration, which yields higher cell viability and enables longer incubation times if needed, and (2) the additional micro-vessel (on the secondary chip), which allows for accurate timing over co-incubation times with the first-in, first-out system. We also acknowledge several disadvantages to this device: (1) The current system does not allow for in-line sorting of successful transconjugants from unconjugated recipients, (2) there is no option for facile, on-chip incubation besides placing the entire device in an incubated chamber, and (3) the current tandem (two-chip) system introduces more potential points of failure, such as leakage between the fluidic tubing connections and the increased potential for air bubble introduction at the tubing-chip interfaces. For example, a downstream reagent inlet stream can be incorporated into the microfluidic platform that allows for the injection of a microdroplet breaking reagent (such as the commercial PicoBreak solution or 1H,1H,2H,2H-perfluoro-1-octanol PFO) to controllably release the inner contents of the droplets. Another possibility is to pair a commercial droplet sorting and plating system (such as the one marketed by On-Chip Biotechnologies or Atrandi Biosciences) to enable a mechanism to detect, sort, and place desired droplets into known areas.
Overall, we envision several areas of application for this device beyond the scope of what is presented in this work, which solely leverages B. subtilis to B. subtilis conjugation. This device can potentially be used with other donor organisms designed for conjugation (e.g., Agrobacterium tumefaciens AGL1) or perhaps even expanded to a tri-parental mating system, such as that described in Banta et al. [12] and Heinze et al. [13]. Moreover, with the addition of upstream and downstream modules, one may also automate the culturing steps leading up to the conjugation as well as the breakage of droplets and subsequent plating steps.
Materials and reagents
Biological materials
Donor strain JAB981 (from Brophy et al. [1]), B. subtilis JH642 Δ(ydcS-yddM)::[(Pspank-GFPmut2) tet(M) cat] thrC::[(int-yddJ) ΔnicK mls] alrA::[(Psweet-rapI) spc]
Recipient strain JAB545 (from Brophy et al. [1]), B. subtilis PY79 cured of ICEBs1, ΔcomC:: aphA-3
Reagents for bacterial culture
LB Miller (Fisher Scientific, catalog number: BP1426-2)
Agar (Fisher Scientific, catalog number: BP1423-500)
Phosphate buffered saline (PBS) (VWR, catalog number: 97062-336)
D-alanine (Sigma-Aldrich, catalog number: A7377)
Tetracycline hydrochloride (Calbiochem, catalog number: 583411; may alternatively use Sigma-Aldrich, catalog number: T7660)
Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: I5502)
NH4Cl (Sigma-Aldrich, catalog number: A9434)
MgCl2 (Sigma-Aldrich, catalog number: M8266)
K2HPO4·3H2O (Sigma-Aldrich, catalog number: P5504)
Materials for chip fabrication and use
SU-8 2075 photoresist (Kayaku, formerly Microchem Corp)
SU-8 2050 photoresist (Kayaku, formerly Microchem Corp)
Microposit EBR 10A (Kayaku, formerly Microchem Corp)
IP-S (Nanoscribe GmbH)
Tridecafluoro-1,1,2,2 tetrahydrooctyl trichlorosilane (United Chemical Technologies LLC, catalog number: T2492)
Poly(dimethylsiloxane) (PDMS) (Dow Corning Corp, PDMS Sylgard 184, catalog number: 761028-5EA)
3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich, catalog number: 440159)
Propylene glycol monomethyl ether acetate (PGMEA) (Sigma-Aldrich, catalog number: 484431)
99% isopropyl alcohol (ULINE, catalog number: S20735)
EBR-10 A (Microposit Kayaku Advanced Materials)
3M Novec 7500 Engineered Fluid (3M, catalog number: 7100025016)
PicoSurf (Sphere Fluidics, catalog number: C021)
Reagents for confirmation assays
Q5 polymerase (NEB, catalog number: M0491L)
dNTPs (NEB, catalog number: N04475)
Q5 reaction buffer (NEB, catalog number: B9027S)
10 μM primers (IDT, exact sequences are noted in the protocol)
Sheath fluid for flow cytometry (Thermo Scientific, NERL Blood Bank Saline, catalog number: 8504)
ReadyLyze Lysozyme (LGC Biosearch Technologies, Lucigen, catalog number: R1804M)
MasterPure Total Nucleic Acid Isolation Kit (LGC Biosearch Technologies, Lucigen, catalog number: MC85200)
Solutions
Tris-Spizizen salts (TSS) buffer (see Recipes)
Carrier (oil) phase for ENTRAP device (see Recipes)
LB agar (see Recipes)
Recipes
Tris-Spizizen salts (TSS) buffer
Reagent Final concentration Quantity or Volume NH4Cl 150 mM 2 g K2HPO4·3H2O 20 mM 0.35 g Tris base (pH 7.5) 49.5 mM 6 g MgCl2 125 mM 11.9 g H2O n/a 1,000 mL Carrier (oil) phase for ENTRAP device
Reagent Final concentration Quantity or Volume PicoSurf (5% in Novec 7500) 1.25% (v/v) 1 mL Novec 7500 n/a 4 mL Total n/a 5 mL LB agar (250 mL)
Reagent Final concentration Quantity or Volume LB 2.5% (w/v) 6.25 g Agar 1.5% (w/v) 3.75 g H2O n/a 250 mL
Laboratory supplies
15 mL culture tubes (Corning, Falcon, catalog number: 352059)
5 mL plastic syringe (BD, catalog number: 309646)
1 mL plastic syringe (BD, catalog number: 309628)
0.2 μm Whatman Puradisc 13 syringe filter (Sigma-Aldrich, catalog number: WHA67801302)
Microtube-130 AFA fiber for sonication (Covaris, catalog number: 520216)
Short Read Eliminator XS kit (Circulomics (PacBio))
Non-DEHP tubing, 30 gauge (Saint-Gobain Performance Plastics, Tygon ND-100-80, catalog number: VWR 89404-300)
Micropipettes 100–1,000 μL (VWR, Eppendorf, catalog number: 89125-306)
Pipette tips 1,000 μL (USA Scientific, TipOne, catalog number: 1122-1830)
96-well microplate (Corning, catalog number: 353072)
Glass slides, 75 × 50 (Corning, catalog number: 2947)
Equipment
Microcentrifuge (Eppendorf, model: Centrifuge 5420)
Shaking incubator (New Brunswick, Innova 42R, catalog number: M1335-0080)
Spectrophotometer (VWR, model: Biowave CO8000 Cell Density Meter)
Stationary incubator (Thermo Fisher Scientific, Heratherm, catalog number: 50125590)
Focused ultrasonicator for genome extractions (Covaris E220 Evolution, catalog number: 500429)
Desiccator (Zoro, catalog number: G4655463)
Spin coater (Laurell Technologies Corporation, model: H6-23)
Photolithography mask aligner (EVG Group, model: EVG 610)
Syringe pump (Chemyx, Fusion 200-X, catalog number: 0720X)
Plasma cleaner (Harrick Plasma, catalog number: PDC-32G)
Optical microscope (Zeiss, model: Colibri Axiovert 200)
Nanoscribe Photonics Professional GT (Nanoscribe GmbH)
Software and datasets
Geneious 2023 (paid software for genetic mapping)
Alternative genetic mapping software: Benchling (free online interface for gene editing)
AutoCAD (paid software for designing master molds)
Alternative CAD software for silicon mold design: Blender, FreeCAD
Filtlong v0.2.1 (https://github.com/rrwick/Filtlong.git)
Raven v1.7.0 (https://github.com/lbcb-sci/raven.git)
medaka v1.4.2 (https://github.com/nanoporetech/medaka.git)
prokka v1.14.5 (https://github.com/tseemann/prokka.git)
Procedure
文章信息
稿件历史记录
提交日期: Jul 6, 2024
接收日期: Sep 8, 2024
在线发布日期: Oct 16, 2024
出版日期: Dec 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Chu, M. J., Wippold, J. A., Renberg, R., Hurley, M., Adams, B. L. and Han, A. (2024). A High-Throughput Droplet-based Method to Facilitate Microbial Conjugation. Bio-protocol 14(23): e5120. DOI: 10.21769/BioProtoc.5120.
分类
生物工程 > 合成生物学 > 基因修饰
生物科学 > 生物技术
微生物学 > 微生物遗传学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link