- EN - English
- CN - 中文
Utilizing the Planar Lipid Bilayer Technique to Investigate Drosophila melanogaster dMpv17 Channel Activity
利用平面脂质双层技术研究果蝇dMpv17通道活性
发布: 2024年11月20日第14卷第22期 DOI: 10.21769/BioProtoc.5118 浏览次数: 1650
评审: Philipp A.M. SchmidpeterAnonymous reviewer(s)
Abstract
The planar lipid bilayer (PLB) technique represents a highly effective method for the study of membrane protein properties in a controlled environment. The PLB method was employed to investigate the role of mitochondrial inner membrane protein 17 (MPV17), whose mutations are associated with a hepatocerebral form of mitochondrial DNA depletion syndrome (MDS). This protocol presents a comprehensive, step-by-step guide to the assembly and utilization of a PLB system. The procedure comprises the formation of a lipid bilayer over an aperture, the reconstitution of the target protein, and the utilization of electrophysiological recording techniques to monitor channel activity. Furthermore, recommendations are provided for optimizing experimental conditions and overcoming common challenges encountered in PLB experiments. Overall, this protocol highlights the versatility of the PLB technique in advancing our understanding of membrane protein function and its broad application in various fields of research.
Key features
• This protocol leverages the planar lipid bilayer (PLB) technique to study the Drosophila Mpv17 protein (WT and mutant forms), revealing its role in forming ion channels.
• The use of a PLB apparatus and advanced electrophysiological recording equipment ensures precise and accurate measurement of ion channel activity.
• The PLB allows for the study of ion channel mechanics and regulation without interference from other proteins, ensuring the purity and accuracy of the results.
Keywords: Electrophysiology (电生理学)Graphical overview
Background
Electrophysiological methods are of pivotal importance in the study of the dynamic functions and physical properties of ion channels, which are essential for cellular signaling and homeostasis [1–5]. Among these methods, the planar lipid bilayer (PLB) technique is held in high regard for its precise analysis of individual ion channel characteristics in a controlled artificial environment [6–8]. This method allows for a detailed investigation of ion channel mechanics at the molecular level and the direct assessment of the effects of various chemicals on these channels, without interference from regulatory proteins present in natural membranes.
The principal advantage of PLB experiments is their capacity to monitor and quantify ion transport across membrane channels, a crucial aspect of cellular physiology. These experiments are conducted in a specially designed chamber comprising two distinct compartments, the cis- and trans-compartments. The aperture between the compartments typically ranges from 50 to 250 μm in diameter, which is necessary for forming a planar bilayer membrane through which lipid-soluble ion channel proteins are integrated. This integration can occur directly from solutions or via fusion with liposomes. Once integrated, the system can measure ionic currents and membrane potentials generated by an electrochemical driving force, providing critical insights into ion conductance and channel activity.
While the PLB method is invaluable for elucidating the intricate relationship between molecular components and physical factors that regulate mammalian ion channels, it does present certain challenges (Table 1). The aperture size can result in prolonged voltage response times and elevated noise levels due to the extensive bilayer area. Nevertheless, contemporary PLB systems with low-noise, high-bandwidth recordings are gradually alleviating these issues, thereby enhancing measurement resolution and precision. Furthermore, the development of sophisticated electrophysiological setups and high-quality synthetic lipids has simplified the formation of artificial membranes and improved reproducibility. However, the variability in membrane proteins, which constitute the ion channels, remains a significant challenge, often determining the success of experimental outcomes.
In a previous study, we investigated the role of the Drosophila melanogaster ortholog of MPV17 (dMpv17) [9]. In humans, mutations in MPV17 are a prominent cause of pediatric-onset hepatocerebral form of mitochondrial DNA depletion syndromes (MDS) (OMIM: 266810) [10–14], including Navajo neurohepatopathy (NHH) [15]. MPV17-dependent MDS is characterized by the rapid deterioration of hepatic function and severe early-onset hypoglycemia, later complicated by neurological impairment. NHH is particularly prevalent in the Navajo population of North America, being caused by the founder R50Q mutation. NHH is characterized by a combination of peripheral neuropathy, liver disease, and other systemic dysfunctions resulting from mitochondrial DNA depletion. The syndrome can manifest in infancy, childhood, or adulthood but is generally associated with longer survival [16]. However, the precise role of MPV17 remains elusive despite decades of intensive research.
Using the planar lipid bilayer (PLB) technique, we investigated the ion channel activity of dMpv17. We showed that dMpv17 exhibited a conductance of 330 pico Siemens (pS) and displayed a preference for cations over anions, like the human counterpart. Furthermore, we demonstrated that the channel facilitated the translocation of uridine but not orotate. Finally, we showed that disease-associated mutations differently impact on channel activity.
This study provides a detailed protocol for PLB setup, expression and reconstitution of ion channel proteins on prepared membranes, ion translocation recordings, and channel current analysis.
Table 1. Key features, benefits, and limitations of PLB. PLB offers customizable membrane compositions and allows the reconstitution of purified proteins and channels, enabling detailed electrophysiological measurements. They provide easy shineaccess to both sides of the bilayer for various assays, making them versatile for a wide range of studies. However, precise control of lipid composition and protein reconstitution can be challenging. PLBs are sensitive to electrical noise and artifacts, and their fragility can lead to experimental failures. Additionally, they require specialized equipment and expertise.
| Feature | Benefits | Limitations |
|---|---|---|
| Membrane composition | Customizable to study various lipid compositions | Requires precise control of lipid composition |
| Reconstitution of proteins | Allows incorporation of purified proteins | Protein reconstitution can be challenging |
| Electrophysiological measurements | Enables detailed ion channel activity analysis | Sensitive to electrical noise and artifacts |
| Accessibility | Easy to access both sides of the bilayer for assays | Bilayer fragility can lead to experimental failure |
| Experimental versatility | Suitable for a wide range of electrophysiological studies | Requires specialized equipment and expertise |
Materials and reagents
L-α-phosphatidylcholine (Sigma-Aldrich, catalog number: P5638)
1,2-diphytanoyl-sn-glycero-3-phosphocholine (4ME 16:0 PC) (Avanti Polar Lipids, catalog number: 850356P)
Decane (Sigma-Aldrich, catalog number: 457116)
Chloroform (Sigma-Aldrich, catalog number: 372978)
Octane (Sigma-Aldrich, catalog number: 296988)
Acetone (Sigma-Aldrich, catalog number: 179124)
Insulin syringes (Biosigma, catalog number: BSS133/A)
Amber glass screw top vials (1.8 mL) (DWK Life Sciences, catalog number: 224750)
Nitrogen stream
Sterile syringe filter with 0.22 μm (Starlab, catalog number: E4780-1226)
Wheat Germ CECF Kit (Biotechrabbit, catalog number: BR1402501)
pIVEX 1.3 WG vector (Biotechrabbit, catalog number: BR1401301)
GenScript SurePAGE, Bis-Tris (GenScript, catalog number: M00658) and Tris-MOPS-SDS running buffer (GenScript, catalog number: M00138)
Bio-Rad Mini-Protean Tetra Cell System
In-Fusion® HD Cloning kit (Takara, catalog number: 639650)
KCl (Sigma-Aldrich, catalog number: P9541)
Tris (Sigma-Aldrich, catalog number: T1503)
KOH (Sigma-Aldrich, catalog number: P5958)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: 05030)
N,N-Dimethyl-n-dodecylamine N-oxide (LDAO) (Sigma-Aldrich, catalog number: 40236)
Uridine (Sigma-Aldrich, catalog number: U3750)
Sodium hypochlorite (Sigma-Aldrich, catalog number: 1056142500)
Primers (Thermo Fisher):
Forward: CCACAACAGCTTGTCGAACCATGAAGAGACTTAAAGCGTA
Reverse: TGATGATGAGAACCCCCCCCGCTATTAAGTATCATGGAGAG
QuickChange II Site-Directed Mutagenesis Kit (Agilent, catalog number: 200523)
R41Q: GAAACGGAGGGTTTGCCCCGCATCCCA and TGGGATGCGGGGCAAACCCTCCGTTTC
S163F: CCCGCTATTAAGTATCATGAAGAGGTAGCAGTTCCATAC and GTATGGAACTGCTACCTCTTCATGATACTTAATAGCGGG
Proteins
WT dMpv17
S163F dMpv17
R41Q dMpv17
Solutions
Bathing solution (pH 7.4) (see Recipes)
10% SDS (see Recipes)
10% LDAO (see Recipes)
1 M KCl (see Recipes)
Uridine (see Recipes)
NaClO (see Recipes)
Recipes
Note: See General note 1.
Bathing solution (pH 7.4)
Reagents Concentration of working solution Amount Final volume Comment KCl 150 mM 1.68 g 150 mL Final volume of solution in the PLB cuvette is 3 mL Tris 10 mM 0.18 g 150 mL KOH 100% Some drops to adjust pH 10% SDS
Reagents Concentration of stock solution Amount Volume of stock solution Comment SDS 10% 1 g 10 mL The final concentration in the experiment is 2% 10% LDAO
Reagents Concentration of stock solution Amount Final volume of stock solution Comment LDAO 10% 1 g 10 mL The final concentration in the experiment is 2% 1 M KCl
Reagents Final concentration Amount Total with the final volume KCl 1 M 37.275 g 500 mL Uridine
Reagents Concentration of stock solution Amount Final volume of stock solution Comment Uridine 200 mM 1.221 g 25 mL The final concentration in the experiment is 10 mM NaClO
Reagents Final concentration Sodium hypochlorite 100%
Equipment
General equipment
50 mL tubes (Corning, catalog number: 430828)
Refrigerated tabletop centrifuge (Hettich, model: MIKRO 220/220R centrifuge)
Vacuum system and vacuum gas pump (VWR Mini Laboratory Pump, VP 86, catalog number: 181-0067P)
Thermomixer (Eppendorf ThermoMixer C, catalog number: EP5382000031)
PLB Equipment (Figure 1)
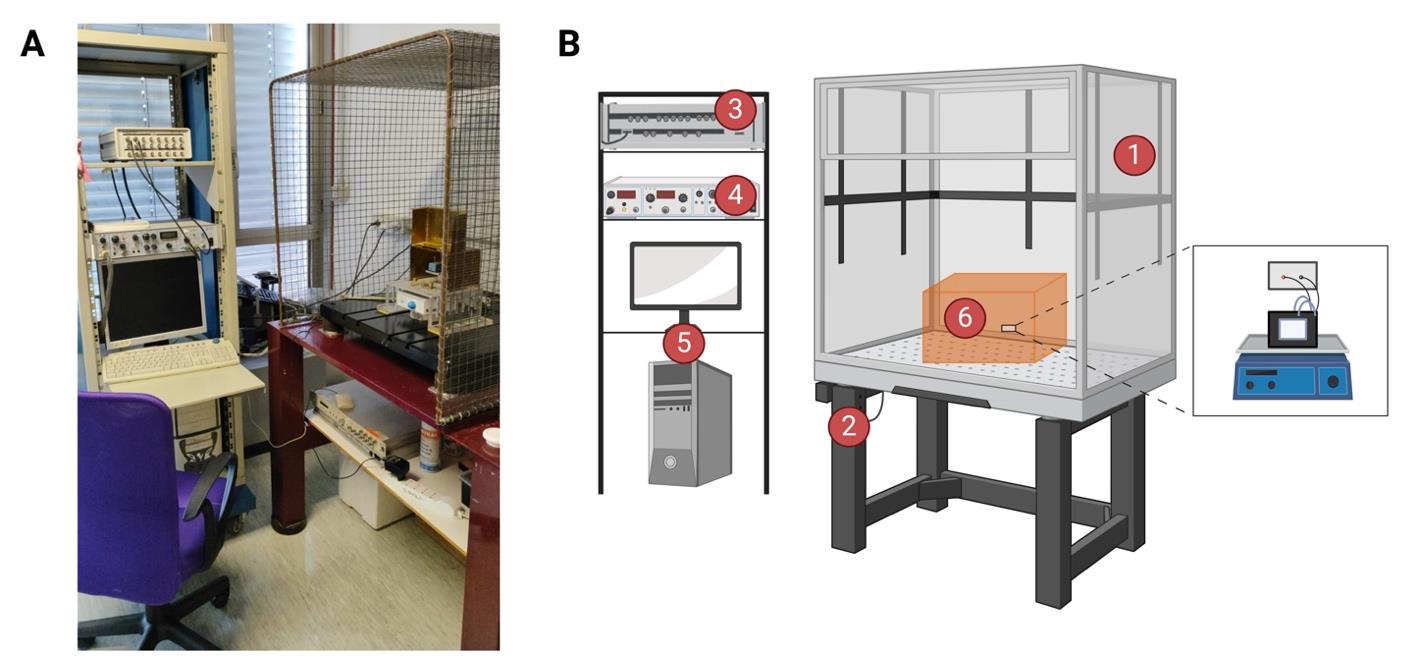
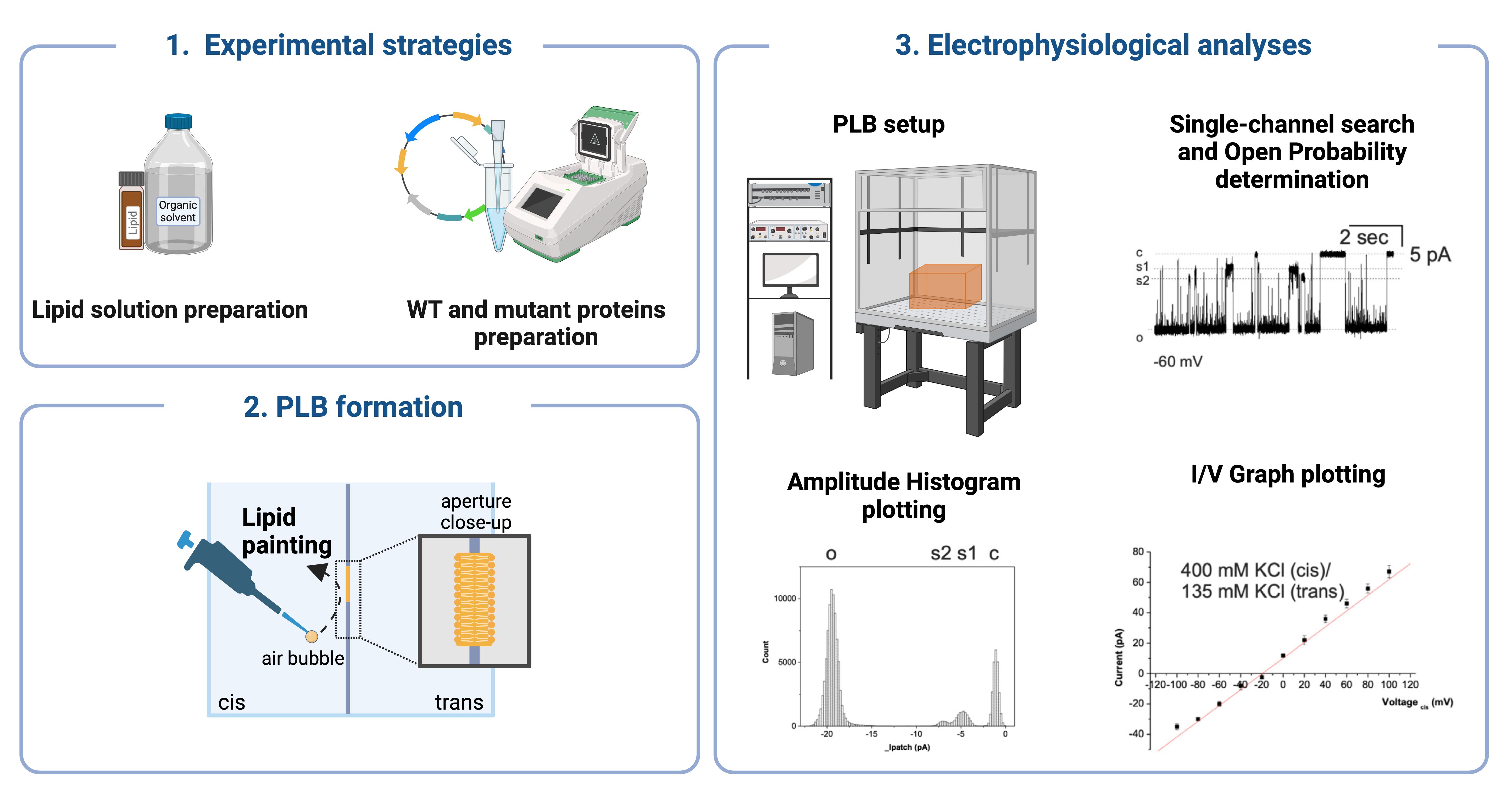
Figure 1. Bilayer workstation. (A) The bilayer workstation used for dMpv17 experiments. (B) Graphical representation of bilayer workstation. The planar lipid bilayer workstation consists of (1) a Faraday cage positioned on an anti-vibration isolation table (2), (3) a bilayer clamp amplifier (e.g., BC-525, Warner Instruments Corporation), (4) a digitizer (e.g., Digidata 1322A, Axon Instruments), a 16-bit device connected to the current and voltage outputs and to a data acquisition system, and (5) a personal computer with an acquisition program (e.g., pCLAMP program sets) for data acquisition. The central part of the workstation is the headstage holder system (6), where the chamber and cuvette system are placed on a magnetic stirrer. Created with BioRender.com.
Faraday cage with vibration isolation table (see General note 2) (Figure 2)
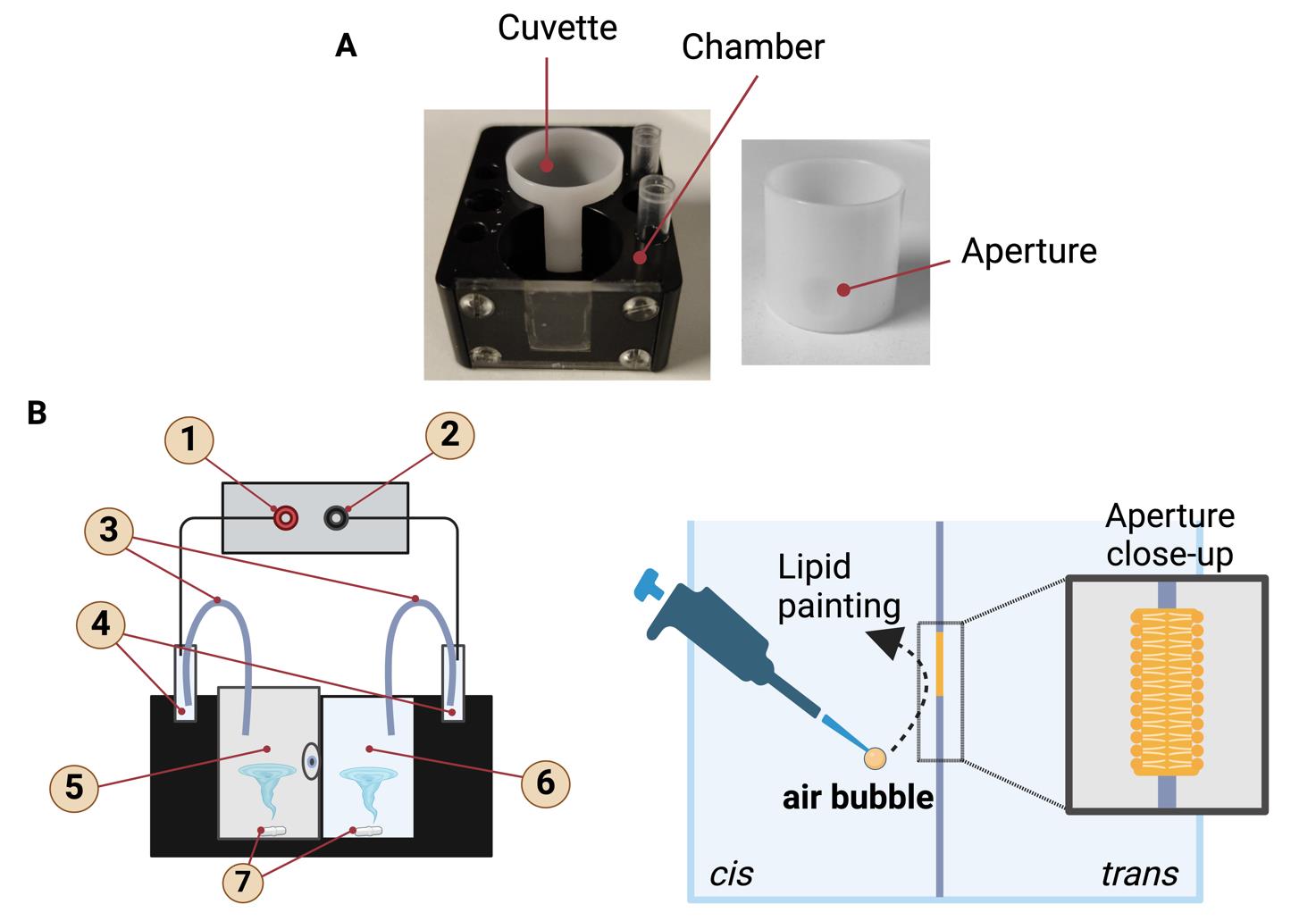
Figure 2. Planar lipid bilayer chamber and cuvette. (A) The lipid bilayer chamber and cuvette, which is inserted into the trans-cavity. The small hole visible on the cuvette is the aperture where the lipid bilayers are formed. Lipid bilayers are painted on the aperture in the center of the cuvette. (B) Graphical representation of the lipid bilayer chamber with the inserted cuvette. The labeled parts are (1) input, (2) reference, (3) salt bridges, (4) Ag-AgCl electrodes, (5) trans side, (6) cis side, (7) magnetic stir bar. Schematic representation of the formation of a lipid bilayer using the "lipid painting" technique. The pipette deposits lipids onto an aperture between two compartments, cis and trans, facilitating the formation of the lipid bilayer. An air bubble aids in the even distribution of the lipids. The close-up shows the structure of the lipid bilayer, with hydrophilic heads facing outward and hydrophobic tails inward. Created with BioRender.com.Bilayer clamp amplifier (Warner Instruments Corporation, model: BC-525)
Digitizer (Axon Instruments, model: Digidata 1322A, 16-bit) (see General note 3)
Bilayer clamp probe (see General note 4) (Figure 2) (Warner Instruments)
Bilayer chamber classic 22 mm chamber (3 mL volume) BCH-M22 (Warner Instruments, catalog number: W4 64-0453)
Classic 22 mm cuvettes Delrin with 250 μm aperture CD22A-250 (Warner Instruments, catalog number: W4 64-0411 (see General note 5)
Homemade salt bridges (see General note 6)
Ag-AgCl electrodes (Warner Instruments, catalog number: WA 10-5, 64-1327)
Magnetic stir bar and magnetic micro stirrer (VELP SCIENTIFICA, catalog number: F203A0161)
5 mm Teflon coated magnets (Warner Instruments, catalog number: 64-0420)
Personal computer equipped with software for data acquisition and analyses
Software and datasets
Clampfit 8 and/or 10.7.0.3 (Molecular Devices LLC)
ORIGIN 6.1 (OriginLab)
Graph Pad Prism 9.0.0 (GraphPad Software LLC)
Procedure
文章信息
稿件历史记录
提交日期: Jul 29, 2024
接收日期: Sep 17, 2024
在线发布日期: Oct 15, 2024
出版日期: Nov 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Corrà, S., Kravenska, Y. and Checchetto, V. (2024). Utilizing the Planar Lipid Bilayer Technique to Investigate Drosophila melanogaster dMpv17 Channel Activity. Bio-protocol 14(22): e5118. DOI: 10.21769/BioProtoc.5118.
分类
生物化学 > 蛋白质 > 活性
生物物理学 > 电生理
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link