- EN - English
- CN - 中文
Multiplex Genome Editing of Human Pluripotent Stem Cells Using Cpf1
利用Cpf1实现人类多能干细胞的多重基因组编辑
发布: 2024年11月20日第14卷第22期 DOI: 10.21769/BioProtoc.5108 浏览次数: 2644
评审: Thirupugal GovindarajanVishal NehruAnonymous reviewer(s)

相关实验方案
利用EpiCRISPR系统通过靶向DNA甲基化诱导Alpha TC1-6细胞产生胰岛素
Marija B. Đorđević [...] Melita S. Vidaković
2025年10月20日 1231 阅读
Abstract
Targeted genome editing of human pluripotent stem cells (hPSCs) is critical for basic and translational research and can be achieved with site-specific endonucleases. Cpf1 (CRISPR from Prevotella and Francisella) is a programmable DNA endonuclease with AT-rich PAM sequences. In this protocol, we describe procedures for using a single vector system to deliver Cpf1 and CRISPR RNA (crRNA) for genome editing in hPSCs. This protocol enables indel formation and homologous recombination–mediated precise editing at multiple loci. With the delivery of Cpf1 and a single U6 promoter-driven guide RNA array composed of an AAVS1-targeting and a MAFB-targeting crRNA array, efficient multiplex genome editing at the AAVS1 (knockin) and MAFB (knockout) loci in hPSCs could be achieved in a single experiment. The edited hPSCs expressed pluripotency markers and could differentiate into neurons in vitro. This system also generated INS reporter hPSCs with a 6 kb cassette knockin at the INS locus. The INS reporter cells can differentiate into β-cells that express tdTomato and luciferase, permitting fluorescence-activated cell sorting of hPSC-β-cells. By targeted screening of potential off-target sequences that are most homologous to crRNA sequences, no off-target mutations were detected in any of the tested sequences. This work provides an efficient and flexible system for precise genome editing in mammalian cells including hPSCs with the benefits of less off-target effects.
Key features
• A single-vector system to deliver Cpf1 and crRNA enables the sorting of transfected cells
• Efficient and simultaneous multi-modular genome editing exemplified by mutation of MAFB and knockin of AAVS1 loci in a single experiment
• Edited PSCs showed minimal off-target effects and can be differentiated into multiple cell types
Keywords: Human pluripotent stem cells (hPSCs) (人类多能干细胞(hPSCs))Graphical overview
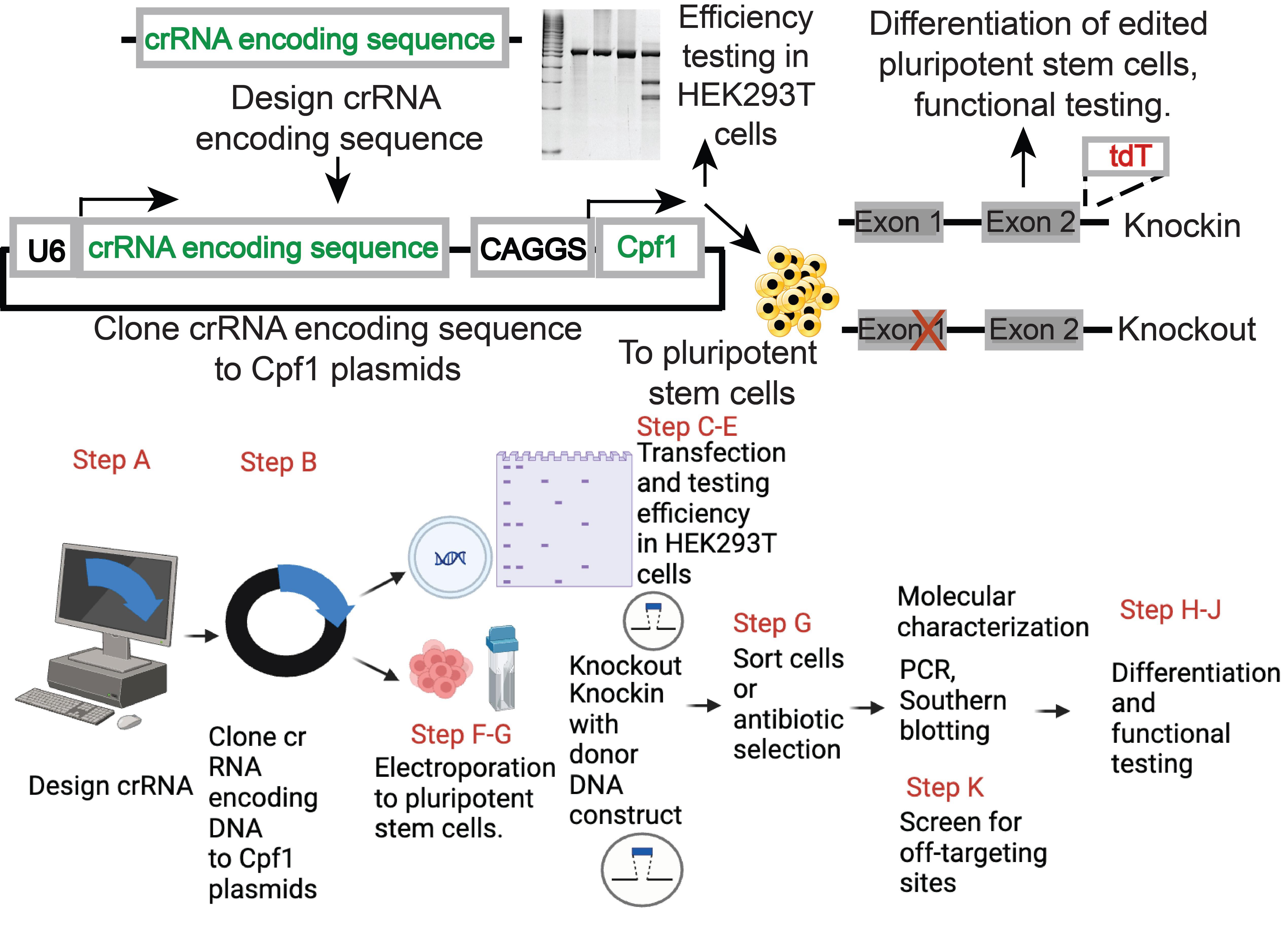
Genome editing of human pluripotent stem cells (hPSCs) using Cpf1. The top panel provides a brief overview of the approach; the bottom panel indicates additional details corresponding to the procedures of the protocol. The bottom panel was generated with biorender.com.
Background
Human pluripotent stem cells (hPSC) [1,2] and their differentiated cell types, such as pancreatic β-cells [3], enable cell replacement therapies as a potential treatment for multiple diseases [4]. Genetic modifications of hPSCs can correct genetic lesions and provide functions including increased immune tolerance by overexpressing PDL1 [5]. Multiple programmable nucleases have been developed for efficient genome editing in hPSCs: zinc finger nucleases [6], TALE nucleases [7], and CRISPR-Cas9 [8,9]. Complementary utilization of these nucleases is a critical component for hPSC-based regenerative medicine [10,11].
The CRISPR-Cas9 system enables efficient genome editing applicable to diverse cell types [12]. However, wild-type SpCas9 can generate double-strand DNA breaks in off-target sites partially complementary to guide RNA. The off-target effects of SpCas9 can be reduced by modifying the basic amino acid residues of the DNA binding domain of SpCas9, but the modified SpCas9 could also show reduced activity compared to wild-type SpCas9 [13]. Base-editing [14] and prime-editing [15] technologies enable genome editing without generating double-stranded DNA breaks. However, introducing large DNA fragments to specific loci of cells remains challenging as recent studies showed insertions in the range of hundreds of nucleotides [16]. Furthermore, to achieve effective multiplex editing at the single-cell level with a DNA vector, each guide RNA requires its promoter, complicating multiplex applications [17].
CRISPR-Cpf1 family endonucleases confer efficient genome editing in transformed mammalian cell lines [18] and mouse embryos [19,20]. Additionally, Francisella novicida Cpf1 (FnCpf1) and Acidaminococcus Cpf1 (AsCpf1) have RNase activity to process their cognate CRISPR RNA (crRNA) [18,21], making it possible to deliver multiple crRNAs from a pre-crRNA array driven by a single U6 promoter [22]. Furthermore, Cpf1 has less off-target effects in genome editing experiments with transformed human cell lines [23,24]. These unique features make Cpf1 a system that complements Cas9.
This protocol describes a detailed genome-editing procedure that uses a single vector system that expresses AsCpf1 and its crRNA. With donor constructs, locus-specific knockin alleles of hPSC were generated and could be differentiated into neurons and β-cells. Furthermore, by introducing a pre-crRNA array composed of AAVS1-targeting guide and MAFB-targeting guide, we observed 100% editing of MAFB locus in AAVS1 targeted clones, providing an example for genome editing in multiple loci in hPSCs. This protocol provides a flexible system for genome editing in hPSCs.
Materials and reagents
Biological materials
H1-OCT4-GFP pluripotent stem cells (WiCell Research Institute, catalog number: H1 OCT4-EGFP, NIH Human Embryonic Stem Cell Registry ID: NIHhESC-10-0043)
WIBR3 pluripotent stem cells (Whitehead Institute, NIH Human Embryonic Stem Cell Registry ID: NIHhESC-10-0079)
HEK293T cells (ATCC, catalog number: CRL-3216)
Stbl3 Chemically Competent E. coli (Invitrogen, catalog number: C737303)
DR4 mice [Dnmt1tm3Jae Hprt1b-m3 Tg(pPWL512hyg)1Ems/J] (Jackson Laboratory, catalog number: 003208)
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory, catalog number: 005557)
DMEM-F12 medium (Life Technologies, catalog number: 11330-057)
KSR (Life Technologies, catalog number: 10828-028)
FGF2 (Life Technologies, catalog number: PHG0261)
2-Mercaptoethanol (2-ME) (Life Technologies, catalog number: 21985-023)
100× L-glutamine (Life Technologies, catalog number: 25030-081)
100× GlutaMAX (Life Technologies, catalog number: 35050-061)
100× MEM-NEAA (Life Technologies, catalog number: 11140-050)
100× penicillin/streptomycin (Life Technologies, catalog number: 15140-122)
100× Insulin-transferrin-selenium-ethanolamine (Life Technologies, catalog number: 51500-056)
Collagenase IV (Life Technologies, catalog number: 17104019)
mTeSR1 medium (STEMCELL Technologies, catalog number: mTeSR1)
Accutase (STEMCELL Technologies, catalog number: 7920)
MCDB131 medium (Life Technologies, catalog number: 10372019)
Y-27632 (Thermo Fisher Scientific, catalog number: 688000-100MG)
Puromycin (Sigma-Aldrich, catalog number: P7255-100MG)
CHIR99021 (Cayman Chemical, catalog number: 252917-06-9)
Matrigel (Thermo Fisher Scientific, catalog number: CB40234)
Activin A (R&D Systems, catalog number: 338-AC-050)
KGF (PeproTech, catalog number: 100-19-250UG)
Vitamin C (Sigma-Aldrich, catalog number: A4544-25G)
Retinoic acid (Sigma-Aldrich, catalog number: R2625-100MG)
SANT-1 (Sigma-Aldrich, catalog number: S4572-5MG)
LDN193189 (Sigma-Aldrich, catalog number: SML0559-5MG)
Heparin (Sigma-Aldrich, catalog number: H3149)
TPB (Tocris, catalog number: 5343)
EGF (PeproTech, catalog number: AF-100-15)
T3 (EMD, catalog number: 64245)
ALK5 inhibitor II (Axxora, catalog number: ALX-270-445-M005)
Gamma secretase inhibitor XX (VWR, catalog number: 82602-300)
Trace Elements A (Corning, catalog number: 25-021-CI)
Trace Elements B (Corning, catalog number: 25-022-CI)
Fatty acid–free BSA (Thermo Fisher Scientific, catalog number: 50412870)
D-Luciferin potassium salt (Perkin Elmer, catalog number: 122799)
RPMI 1640 (Gibco, catalog number: 31800-089)
DMEM high glucose medium (Cytiva, catalog number: SH30022)
FBS (Cytiva, catalog number: SH30396.03HI)
BbsI-HF (New England Biolabs, catalog number: R3539S)
SphI-HF (New England Biolabs, catalog number: R3182L)
CutSmart Buffer (New England Biolabs, catalog number: B6004)
T7 Endonuclease I (New England Biolabs, catalog number: M0302S)
Lipofectamine 2000 transfection reagent (Invitrogen, catalog number: 11668019)
Plasmid Mini Kit (Omega Bio-Tek, catalog number: D6943-02)
DNA Clean & Concentrator (Zymo Research, catalog number: D4033)
T4 PNK (New England Biolabs, catalog number: M0201S)
T4 DNA ligase (New England Biolabs, catalog number: M0202M)
DNeasy Blood & Tissue Kit (Qiagen, catalog number: 69504)
Q5 HiFi 2× Master Mix (New England Biolabs, catalog number: M0492L)
10% TBE PAGE gel (Bio-Rad, catalog number: 3450053)
Mouse anti-OCT3/4 antibody, 1:100 (BD Transduction Laboratories, catalog number: 611203)
Rabbit anti-NANOG antibody, 1:100 (Thermo Fisher Scientific, catalog number: PA1-097)
Mouse anti-TRA-1-60, 1: 100 (Life Technologies, catalog number: 411000)
Mouse anti-SSEA4 antibody, clone MC-813-70, 1:100 (Thermo Fisher Scientific, catalog number: MA1-021)
Mouse anti-Nestin antibody, 1:100 (Santa Cruz, catalog number: SC-23927)
Mouse anti-TUBB3 antibody, 1:1000 (BioLegend, catalog number: 801201)
Alexa 488 conjugated anti-mouse IgG antibodies, used at 1:400 (Life Technologies, catalog number: A21202)
Alexa 488 conjugated anti-rabbit IgG antibodies, used at 1:400 (Life Technologies, catalog number: A21206)
ZymoPURE II Plasmid Midiprep Kit (Zymo Research, catalog number: D4200)
AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific, catalog number: 4398881)
E.Z.N.A.® Gel Extraction kit (Omega Bio-Tek, catalog number: D2500-02)
HindIII-digested Lamda DNA (New England Biolabs, catalog number: N3012S)
Pyruvate (100 mM) (Corning, catalog number: 25-000-CIR)
Insulin solution (Sigma, catalog number: I9278)
Neurobasal medium (Life Technologies, catalog number: 21103049)
AlbuMAX I Lipid-Rich BSA (Life Technologies, catalog number: 11020021)
N2 NeuroPlex (GeminiBio, catalog number: 400163)
Lactic syrup (Sigma, catalog number: L4263-100ML)
Biotin (Sigma, catalog number: B4639-500MG)
Gem21 without vitamin A (GeminiBio, catalog number: 400-161-010)
Proteinase K (Promega, catalog number: C3021)
Sodium citrate tribasic dihydrate (Sigma, catalog number: C8532)
Primers
F NGS seq primer for MAFB of WIBR3 602-1 ATATGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-2 AGAGGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-3 ACACGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-4 GCGCGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-5 GTGTGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-6 CTCTGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-7 TTAAGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-8 TTGGGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-9 TTCCGGTCAAGTGCGAGAAACTCG
F NGS seq primer for MAFB of WIBR3 602-10 CCTTGGTCAAGTGCGAGAAACTCG
R NGS seq primer for MAFB of WIBR3 GCAGGGACAGGGTCCGGGGTAG
AAVS1 off 1 F CTGCTGAACACTCAGCATCTGCC
AAVS1 off 1 R CAGAGGAGCGAGTGGAGCAGACAG
AAVS1 off 2 F ATTGCAATATCCTCCTATTAGCC
AAVS1 off 2 R CACTAGAGTCACCCTATGGCTCCC
AAVS1 off 3 F CATGGGACAAGTTGATAGCTAAG
AAVS1 off 3 R GAAGTTACTCTGAAACGTATAGCAC
AAVS1 off 4 F GCTGCTCCTGGATTTAGCAAAC
AAVS1 off 4 R GCCCAGCCCAACACTTTTGGTC
AAVS1 off 5 F GTGTGCTAGTATCATTGCAAAAG
AAVS1 off 5 R CACTATTGCGTTCTCTCATTTCTC
MAFB off 1 F TCTGAGGTCCGTCTCACACACTG
MAFB off 1 R CACTGTTCAAAGAGTTTGAACATTCC
MAFB off 1 F CTCCTGACTATTGCAGTTGCTGGTCACC
MAFB off 1 R AACAGAGGAGCGAGTGGAGC
INS off 1 F CGGAGTCTCACTTTGTTGCCATG
INS off 1 R TGGTTAGAACTTCCTGCCCACAG
Gelatin (Sigma, catalog number: G2500)
Mitomycin C (Sigma, catalog number: M4287)
CMV-AsCpf1-2A-GFP-U6-AAVS1-crRNA plasmid (Addgene, #194716)
AVS1-tdT targeting plasmid (Addgene, #194728)
CAGGS-AsCpf1-2A-GFP-U6-crRNA-cloning vector (Addgene, #159281)
CMV:AsCpf1-2A-GFP-U6-crRNA-cloning vector (Addgene, #194715)
CAGGS-AsCpf1-2A-GFP-U6-tdT-crRNA plasmid (Addgene, #194724)
CAGGS-AsCpf1-2A-GFP-U6-AAVS1-crRNA plasmid (Addgene, #194723)
CAGGS-AsCpf1-2A-GFP-U6-AAVS1-MAFB-crRNA (Addgene, #194725)
CAGGS-AsCpf1-2A-GFP-U6-INS-crRNA plasmid (Addgene, #159283)
INS-2A-luciferase-2A-tdT donor plasmid (Addgene, #159348)
LB broth (Sigma, catalog number: L3522-1KG)
Agar (Fisher Scientific, catalog number: 1423-500)
Na2HPO4·7H2O (Sigma, catalog number: S9390)
Na2HPO4 (Fisher Scientific, catalog number: BP329-500)
NaCl (Sigma, catalog number: S271-3)
KCl (Fisher Scientific, catalog number: BP366-1)
KH2PO4 (Sigma, catalog number: P0662)
CaCl2 (Sigma, catalog number: C8106-500G)
MgCl2·6H2O (Sigma, catalog number: M2670-500G)
Tris (Millipore, catalog number: 648311-1KG)
EDTA (Sigma, catalog number: E6758-500g)
SDS (Fisher Scientific, catalog number: BP166-500)
H3PO4 (85%) (Sigma, catalog number: 345245-500ML)
Ascorbic acid (Sigma, catalog number: A4544-25G)
REDTaq PCR Reaction Mix (Sigma, catalog number: R2523)
Prime-It II Random Primer Labeling Kit (Agilent, catalog number: 300385)
CHROMA SPIN + TE-30 (Takarabio, catalog number: 636093)
Hybond-XL membrane (Cytiva, catalog number: RPN2020S)
BioMax MS film (Carestream, catalog number: 8294958)
NaOH (Sigma, catalog number: S5881-500G)
HCl (Sigma, catalog number: H1758)
Dorsomorphin (R & D Systems, catalog number: 3093/10)
Acetic acid (Sigma, catalog number: A6283)
Ethidium bromide (Sigma, catalog number: E1510)
[a-32P]dCTP (Revvity, catalog number: BLU513H)
LB agar (see Recipes)
Gelatin solution (see Recipes)
10× PBS (see Recipes)
Ca2+ and Mg2+ solution (see Recipes)
PBS+ (see Recipes)
MEF medium/HEK293T cells medium (see Recipes)
MEF inactivation medium (see Recipes)
DNA-extraction buffer (see Recipes)
2× phosphate buffer (pH 7.2) (see Recipes)
Hybridization buffer (see Recipes)
20× SSC buffer (pH 7.0) (see Recipes)
hPSC medium (see Recipes)
NGD medium (see Recipes)
50× TAE buffer (see Recipes)
LB agar
Reagent Final concentration Quantity or Volume LB broth 2.5% w/vol 10 g Agar 1.5% w/vol 6 g MilliQ water n/a Add to 400 mL Total (optional) n/a 400 mL Gelatin solution
Reagent Final concentration Quantity or Volume Gelatin 0.2% (w/v) 0.8 g H2O n/a 400 mL Total n/a 400 mL 10× PBS
Reagent Final concentration Quantity or Volume Na2HPO4·7H2O 25.6 g Na2HPO4 13.56 g NaCl 80 g KCl 2 g KH2PO4 2 g H2O To 1,000 mL Total 1,000 mL Adjust pH to 7.4 and autoclave for 15 min at 121 °C.
Ca2+ and Mg2+ solution
Reagent Final concentration Quantity or Volume CaCl2 5 g MgCl2·6H2O 5 g H2O To 500 mL Total 500 mL Filter the solution through a 0.22 μm filter and autoclave for a 45 min liquid cycle. After autoclaving, keep the solution at 4 °C.
PBS+
Reagent Final concentration Quantity or Volume 1× PBS (diluted from 10× PBS) 990 mL Ca2+ and Mg2+ solution (Recipe 4) 10 mL Total 1000 mL Stir the solution for approximately 30 min until it clears up. Then, filter through a 0.22 μm filter and keep refrigerated at 4 °C.
MEF medium/HEK293T cells medium
Reagent Final concentration Quantity or Volume DMEM high glucose 435 mL FBS 10% 50 mL Penicillin/streptomycin 1% 5 mL NEAA 1% 5 mL Pyruvate (100 mM) 1 mM 5 mL Total 500 mL MEF inactivation medium
Reagent Final concentration Quantity or Volume DMEM high glucose 87 mL FBS 10% 10 mL Penicillin/streptomycin 1% 1 mL NEAA 1% 1 mL Pyruvate (100 mM) 1 mM 1 mL Mitomycin C 5 µg/mL 0.5 mg Total 100 mL DNA-extraction buffer
Reagent Final concentration Quantity or Volume 1 M Tris (pH 8.0) 50 mM 25 mL 0.5 M EDTA (pH 8.0) 10 mM 10 mL 5 M NaCl 100 mM 10 mL 10% SDS 0.5% 25 mL MilliQ H2O 430 mL 20 mg/mL Proteinase K 0.5 mg/mL 25 μL for 1 mL, add just before use 2× phosphate buffer (pH 7.2)
Reagent Final concentration Quantity or Volume Na2HPO4·7H2O 0.5 M 67 g H3PO4 (85%) 4 mL MilliQ H2O Add to 1 L Hybridization buffer
Reagent Final concentration Quantity or Volume 2× phosphate buffer (Recipe 9) 1× 50 mL 0.5 M EDTA (pH 8.0) 1 mM 0.2 mL 5 M NaCl 100 mM 5 mL 20% SDS 7 % 35 mL MilliQ H2O 15 mL BSA 1% 1 g, add before use in 100 mL buffer warmed to 60 °C 20× SSC buffer (pH 7.0)
Reagent Final concentration Quantity or Volume NaCl 175.3 g Sodium citrate tribasic dihydrate 88.2 g MilliQ H2O Add to 1 L, adjust pH to 7.0 with a few drops of HCl hPSC medium
Reagent Final concentration Quantity or Volume DMEM-F12 385 mL KSR 5% 25 mL FBS 15% 75 mL Penicillin/streptomycin 1% 5 mL NEAA 1% 5 mL L-glutamine 1% 5 mL FGF2 4 ng/mL 2-ME 1 μM Total (optional) n/a 500 mL NGD medium
Reagent Final concentration Quantity or Volume Neurobasal medium 500 mL Gem21 without vitamin A 1% 5 mL AlbuMAX I 0.2% w/v 1 g NeuroPlex N2 0.5% 2.5 mL NaCl (5 M) 5 mL Pyruvate (100 mM) 1 mM 5 mL Penicillin/streptomycin (100×) 1× 5 mL GlutaMAX (100×) 1× 5 mL Biotin (5 mg/mL in 1 M NaOH) 0.35 μL Ascorbic acid (100 mM) 50 μL Lactic syrup (85%) 100 μL 50× TAE
Reagent Final concentration Quantity or Volume Tris 2 M 121 g EDTA (0.5 M, pH 8) 50 mM 50 mL Acetic acid 1 M 28.55 mL H2O To 500 mL Total 500 mL
Laboratory supplies
6-well ultra-low attachment plates (Corning, catalog number: CLS3471-24EA)
AggreWell 400 (StemCell Technologies, catalog number: 34425)
12-well plates (Corning, catalog number: 3512)
6-well plates (Corning, catalog number: 3506)
Cell lifters (Corning, catalog number: 3008)
Electroporation cuvettes, 0.2 cm (Bio-Rad, catalog number: 1652086)
PCR tubes and caps (USA Scientific, catalog number: 1402-2500)
Equipment
SMZ1270 microscope (Nikon, model: SMZ1270)
Eclipse Ti microscope (Nikon, model: Eclipse Ti)
Thermal Cycler (Bio-Rad, model: T100)
Accuspin micro desktop centrifuge (Fisher Scientific, catalog number: 75002461)
ST1 Plus desktop centrifuge (Sorvall ST1 Plus, with =TX-400 rotor and adaptors, catalog number: 75016030)
Heracell 150i CO2 incubator (Thermo Scientific, catalog number: 13998034)
Incushaker (Benchmark Scientific, catalog number: 31-206)
Gene Pulser Xcell Eukaryotic System (Bio-Rad, catalog number: 1652661)
Nanodrop 2000 spectrometer (Thermo Scientific, catalog number: ND2000)
AlphaImager gel documentation system (Alpha Innotech, model: AlphaImager 2200)
Xenogen IVIS Spectrum imager (Xenogen, model: IVIS Spectrum)
FACSAria cell sorter (BD Biosciences)
Software and datasets
Cpf1 crRNA design was performed at https://portals.broadinstitute.org/gppx/crispick/public
Prediction of off-targets was obtained using Cas-OFFinder, http://www.rgenome.net/cas-offinder
Procedure
文章信息
稿件历史记录
提交日期: Jun 27, 2024
接收日期: Sep 6, 2024
在线发布日期: Oct 15, 2024
出版日期: Nov 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Ma, H. (2024). Multiplex Genome Editing of Human Pluripotent Stem Cells Using Cpf1. Bio-protocol 14(22): e5108. DOI: 10.21769/BioProtoc.5108.
分类
干细胞 > 多能干细胞 > 再生医学
细胞生物学 > 细胞工程 > CRISPR-cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link