- EN - English
- CN - 中文
PEPITA: Parallelized High-Throughput Quantification of Ototoxicity and Otoprotection in Zebrafish Larvae
PEPITA:斑马鱼幼鱼耳毒性和耳保护性高通量并行定量
发布: 2024年11月05日第14卷第21期 DOI: 10.21769/BioProtoc.5105 浏览次数: 1681
评审: Ivonne SehringAlberto RissoneAnonymous reviewer(s)
Abstract
Drug-induced hearing injury (ototoxicity) is a common, debilitating side effect of many antibiotic regimens that can be worsened by adverse drug interactions. Such adverse drug interactions are often not detected until after drugs are already on the market because of the difficulty of measuring all possible drug combinations. While in vivo mammalian assays to screen for ototoxic damage exist, they are currently time-consuming, costly, and limited in throughput, which limits their utility in assessing drug interaction outcomes. To facilitate more rapid quantification of ototoxicity and assessment of adverse drug interactions that impact ototoxicity, we have developed a high-throughput workflow we call parallelized evaluation of protection and injury for toxicity assessment (PEPITA). PEPITA uses zebrafish larvae to quantify ototoxic damage and protection. Previous work has shown that hair cells (HCs) in the zebrafish lateral line are very similar to human inner ear HCs, meaning zebrafish are a viable model to test drug-induced ototoxicity. In PEPITA, we expose zebrafish larvae to different combinations of drugs, fluorescently label the HCs, and subsequently use microscopy to quantify the brightness of the fluorescently labeled HCs as an assay for ototoxic damage and hair-cell viability. PEPITA is a reproducible, low-cost, technically accessible, and high-throughput assay. These advantages allow many experiments to be conducted in parallel, paving the way for systematic evaluation of drug-induced hearing injury and other multidrug interactions.
Key features
• Analysis of drug-induced hair cell damage associated with ototoxicity using Danio rerio
• Ototoxicity assessment performed in vivo
• Uses microscopy to generate images to assay ototoxicity quantitatively
• Enables testing of various combinations of drugs at various doses to determine toxicity-associated drug–drug interaction outcomes (synergy, antagonism)
Keywords: Zebrafish (斑马鱼)Graphical overview
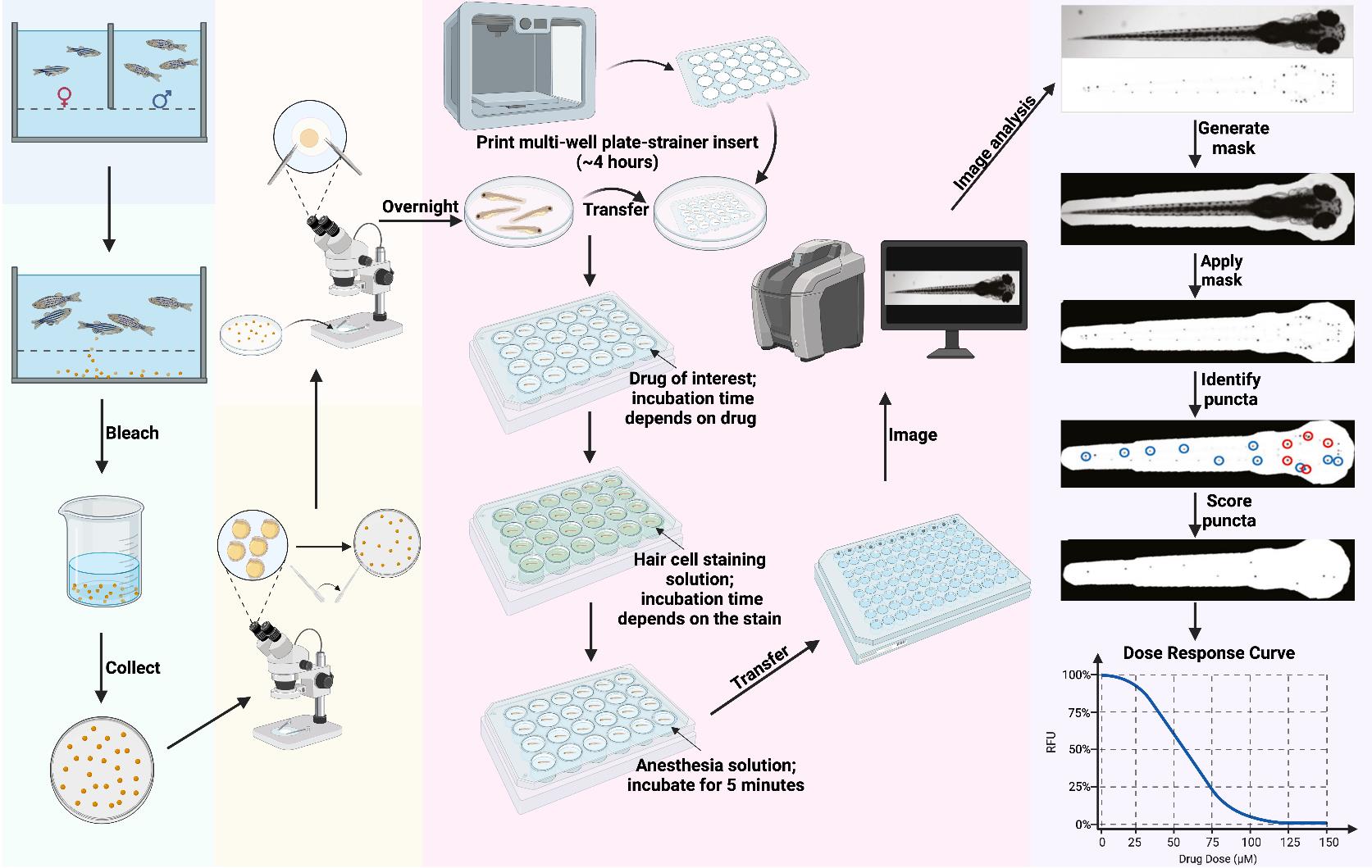
Overview of PEPITA workflow for fish handling. Day 1 (day before fertilization): Set up crossing and incubate overnight. Day 0 (day of fertilization): Execute crossing, bleach the collected embryos for 10 min, transfer embryos into Petri dishes, and incubate overnight. Day 1 (1 day post-fertilization (dpf)): Transfer viable embryos into new Petri dishes under a microscope and incubate overnight. Day 3–4 dpf: Dechorionate the larvae and incubate overnight. Day 5 dpf: Place a 3D-printed multi-well plate-strainer insert into a large Petri dish and transfer the larvae into each well of the insert. Transfer the larvae-containing insert to its respective drug treatment plate for the desired drug exposure time (this varies depending on the drug and the goal of the experiment; we have used treatment durations of 1 min to 24 h). Following this, transfer the inserts into a plate with hair cell staining solution and incubate (this varies depending on the type of stain; we have used staining durations of 1–20 min), then transfer them into a plate with anesthesia solution and incubate until larvae stop moving. Then, transfer the larvae to a 96-well imaging plate and image whole fish in brightfield and relevant fluorescence channels. Imaging time depends on the instrument and settings; our imaging workflow typically takes ~30 min to image a single 96-well plate. Data analysis: The PEPITA software package accepts brightfield (top) and fluorescence (bottom) images, creates and applies a mask to each fluorescence image to focus analysis on the fish. The package next identifies the top 15 brightest local maxima and creates a second mask around ten puncta (blue circles). Then, it applies the second mask and automatically scores each image according to the relative fluorescence intensity of the identified 10 puncta. Relative fluorescence intensities scored from drug-exposed fish can be compared quantitatively to untreated fish to calculate individual dose-response curves or pairwise drug interaction outcome reports. Figure generated in BioRender.
Background
Hearing loss is one of the most common chronic diseases, affecting approximately 30% of individuals aged 65–74 years old and 50% of individuals more than 75 years old (both hearing and balance disorders) [1]. Drug-induced injury to hearing and balance (ototoxicity) is a significant concern, with over 150 medications associated with ototoxicity [2]. Adverse drug–drug interactions (DDIs) pose a challenge in drug treatment, as interactions among medications can amplify the potential for hearing damage. New approaches for preclinical drug screening to assess ototoxicity and identify DDIs are needed for developing safer therapeutic options.
Zebrafish are a well-established vertebrate model for high-throughput profiling and studying ototoxicity. While their tissues and organs share many similarities with those of humans, zebrafish grow rapidly, produce hundreds of embryos per spawn, and are optically transparent as larvae, making them a powerful preclinical tool. Zebrafish have been utilized to assess drug toxicity in many organs and tissues, including cardiovascular, gastrointestinal, and mechanosensory systems [3]. Within the mechanosensory system, zebrafish possess the lateral line, which contains hair cells (HCs) that are homologous to the sensory HCs found in the inner ear of humans. Previous work has demonstrated that ototoxic drugs (e.g., aminoglycoside antibiotics and chemotherapeutics) induce similar HC damage and death in both zebrafish lateral lines and human inner ears [4,5]. Established lateral line damage quantification methods involve confocal microscopy of individual HC clusters (i.e., neuromasts) and manual HC counting. While these approaches provide subcellular resolution on damage phenotypes, they are effort-intensive and thus limited in the number of neuromasts, fish, and conditions that can be profiled per experiment. DDIs quantification would not be tractable with standard neuromast-centric characterization methods.
Here, we describe a high-throughput, semi-automated method for drug screening to assess ototoxicity in vivo using zebrafish, named parallelized evaluation of protection and injury for toxicity assessment (PEPITA). This method is tailored for large-scale drug screening and investigation of DDIs in zebrafish larvae. PEPITA enables efficient quantification of lateral line phenotypes using whole-fish fluorescence microscopy of hundreds of fish per day. Fish can be imaged in multi-well plates after being treated with multiple drugs and fluorescent dyes that report on the phenotype of stained HCs. For example, to measure HC viability, we have used the fluorescent stain oxazole yellow with propidium iodide (YO-PRO-1). Because YO-PRO-1 can only enter through HCs with functioning mechanotransduction channels, it gives a qualitative proxy for HC viability [6,7]. Alternatively, myo6b::gfp fish (transgenic fish that express GFP in their HCs) can be used in place of YO-PRO-1 stained fish as a more cost-effective option for assessing ototoxic damage that also enables longitudinal assessment. PEPITA also includes a software package for quantitatively analyzing images to study the dose-response and DDIs characteristics of individual and pairwise treatment conditions based on the neuromast-specific fluorescence intensity of chemical stains or transgenically expressed fluorescent proteins. Collectively, PEPITA enables semi-automated quantification of lateral line phenotypes measurable with fluorescence readouts. The workflow is amenable to simultaneous quantification of multiple fluorescence channels, and the underlying image analysis pipeline is amenable to adaptation for quantifying toxicity in other zebrafish organs and tissues with fluorescence-based assays.
Materials and reagents
Biological materials
AB strain zebrafish (Danio rerio) (Zebrafish International Resource Center, European Zebrafish Resource Center, China Zebrafish Resource Center)
Tg
; myo6b::gfp zebrafish [8]
Reagents
Note: Azithromycin and neomycin are listed as examples of antibiotics used, but any drugs of interest can be used here.
Azithromycin dihydrate (Thermo Fisher, catalog number: AAJ6674006) (store at 4 °C)
Neomycin sulfate hydrate (Thermo Fisher, catalog number: AAJ6149914)
Texas red-X succinimidyl ester (Thermo Fisher, catalog number: T20175) (store at –20 °C)
Tricaine (3-aminobenzoic acid ethyl ester, methanesulfonate salt) (Pentair, catalog number: TRS5)
YO-PRO-1 Iodide (Thermo Fisher, catalog number: Y3603) (store at –20 °C)
Instant Ocean (Spectrum Brands Product, catalog number: SS15-10)
Bleach (Clorox, catalog number: 30966)
Reverse osmosis (RO) filtered water
Antibiotic super-concentrated stock solution (see Recipes for neomycin example)
YO-PRO-1 stain solution (2 μM) (see Recipes)
Tricaine: stock solution (15 mM) (see Recipes)
Tricaine: euthanasia solution (1.35 mM) (see Recipes)
Tricaine: anesthesia solution (0.675 mM) (see Recipes)
ICS water (see Recipes)
Neomycin super-concentrated stock solution
Note: This is an example of an antibiotic solution. The super-concentrated stock solution concentrations and solvents used will vary between drugs. Store at -20 °C or -80 °C.
Reagent Final concentration Quantity or Volume Neomycin sulfate hydrate 200 mM 36 mg Molecular-grade water 200 μL Total 200 μL YO-PRO-1 solution (2 μM)
Note: The recipe is for one 12-well plate, using 3,500 μL of YO-PRO-1 solution per well. Adjust volumes as needed, depending on the size of the multi-well plate being used. Store at -20 °C.
Reagent Stock concentration Final concentration Quantity or Volume YO-PRO-1 Iodide 1 mM 2 μM 84 μL ICS water (Recipe 6) N/A 42 mL Total 42 mL Tricaine: stock solution (15 mM)
Reagent Stock concentration Final concentration Quantity or Volume Tricaine powder N/A 15 mM 400 mg Nano-pure water N/A 97.9 mL 1 M Tris or 1 M sodium bicarbonate 1 M 21 mM 2.1 mL HCl as needed to reach pH of ~7 Total ~100 mL Store at -20 °C.
Tricaine: euthanasia solution (1.35 mM)
Reagent Stock concentration Final concentration Quantity or Volume Tricaine: stock solution (Recipe 3) 15 mM 1.35 mM 4.5 mL ICS water (Recipe 6) N/A 45.5 mL Total 50 mL Tricaine: anesthesia solution (0.675 mM)
Note: The recipe is for one 12-well plate, using 3,500 μL of tricaine solution per well. Adjust volumes as needed, depending on the size of the multi-well plate being used.
Reagent Stock concentration Final concentration Quantity or Volume Tricaine: euthanasia solution (Recipe 4) 1.35 mM 0.675 mM 25 mL ICS water (Recipe 6) N/A 25 mL Total 50 mL ICS water
Note: Any type of “fish water” or embryo medium safe for use with zebrafish embryos and larvae can be used in place of ICS water. However, bear in mind that differing water composition may cause hair cell sensitivity to damage to vary.
Reagent Final concentration Quantity or Volume Instant Ocean 0.25 g/L 5.0 g Calcium chloride 0.05 g/L 1.0 g Sodium bicarbonate 0.07 g/L 1.3 g Reverse osmosis (RO) filtered water 20 L Total (optional) 20 L
Laboratory supplies
Note: Supplies indicated with an asterisk are brand-specific because the Tinkercad design for the multi-well plate-strainer inserts may not fit multi-well plates from other manufacturers.
Breeding tanks with dividers (Carolina Biological Supply, catalog number: 161937)
12-well plates, non-treated* (Corning Costar, catalog number: 07-201-589)
24-well plates, non-treated* (Corning Costar, catalog number: 07-201-590)
96-well plates, non-treated, black, flat-bottom, for imaging (Thermo Fisher, catalog number: 37000-550)
3D printer filament, PLA (Amazon, catalog number: B084XS2TS9)
Nylon mesh, 125 micron (ELKO Filtering Co., catalog number: 03-125/39)
Wide-bore Pasteur pipettes* (Fisherbrand, catalog number: 13-678-30)
Note: Wide-bore Pasteur pipettes are necessary because the embryos will not fit into standard Pasteur pipets.
100 μm cell strainers (Corning, catalog number: 07-201-432)
Petri dishes (100 mm × 15 mm) (Falcon, catalog number: 08-757-100D)
Large Petri dishes (150 mm × 15 mm) (VWR, catalog number: 25384-326)
Compressed air duster cleaner (Inovera, catalog number: IVR10012)
Fine-tipped tweezers (Dumont Tweezer, Style 5, catalog number: 0203-5-PO)
Equipment
All-in-one fluorescence microscope (Keyence, model: BZ-X710)
3D printer (Dremel, model: Digilab 3D45 3-D printer)
Hot plate (Fisher Scientific, Thermix Stirring Hot Plate, model: 210T)
Vortex mixer (VWR, catalog number: 10153-838)
Digital dry bath (BioRad, catalog number: 1160562)
Software and datasets
Tinkercad design for multi-well plate-strainer inserts (freely available at https://www.thingiverse.com/thing:5676024)
PEPITA-tools software package can be found on GitHub at https://github.com/ma-lab-cgidr/PEPITA-tools
ImageJ (version 1.54f, 06/29/2023)
Procedure
文章信息
稿件历史记录
提交日期: Jul 14, 2024
接收日期: Sep 13, 2024
在线发布日期: Oct 12, 2024
出版日期: Nov 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Nilles, E. M., Bustad, E., Qin, M., Mudrock, E., Gu, A., Galitan, L., Ou, H. C., Hernandez, R. E. and Ma, S. (2024). PEPITA: Parallelized High-Throughput Quantification of Ototoxicity and Otoprotection in Zebrafish Larvae. Bio-protocol 14(21): e5105. DOI: 10.21769/BioProtoc.5105.
分类
生物科学 > 生物技术
药物发现
细胞生物学 > 细胞成像 > 活细胞成像
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link