- EN - English
- CN - 中文
Single Cell Isolation from Human Diabetic Fibrovascular Membranes for Single-Cell RNA Sequencing
从人类糖尿病纤维血管膜中分离单细胞用于单细胞RNA测序
发布: 2024年10月20日第14卷第20期 DOI: 10.21769/BioProtoc.5096 浏览次数: 1909
评审: Pilar Villacampa AlcubierreYu LiuPreeti Yadav
Abstract
Single-cell transcriptomic analyses have emerged as very powerful tools to query the gene expression changes at the single-cell level in physiological and pathological conditions. The quality of the analysis is heavily dependent on tissue digestion protocols, with the goal of preserving thousands of single live cells to submit to the subsequent processing steps and analysis. Multiple digestion protocols that use different enzymes to digest the tissues have been described. Harsh digestion can damage certain cell types, but this might be required to digest especially fibrotic tissue as in our experimental condition. In this paper, we summarize a collagenase type I digestion protocol for preparing the single-cell suspension from fibrovascular tissues surgically removed from patients with proliferative diabetic retinopathy (PDR) for single-cell RNA sequencing (scRNA-Seq) analyses. We also provide a detailed description of the data analysis that we implemented in a previously published study.
Key features
• Single-cell suspension from fibrovascular membranes isolated from PDR patients.
• Single-cell RNA sequencing analyses performed using Seurat package in RStudio.
• Trajectory analyses or pseudotime analyses to study the trajectory over (pseudo)time of specific cell types.
• This protocol requires Illumina HiSEQ4000 instrument and knowledge of R and RStudio language for the analyses.
Keywords: scRNA-Seq (单细胞RNA测序)Graphical overview
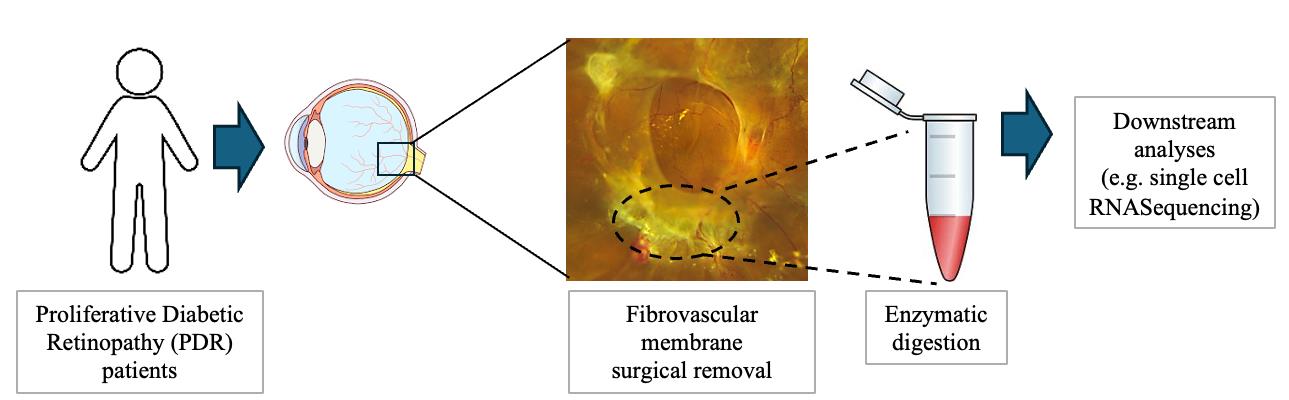
Fibrovascular membrane isolation and sequencing
Background
Proliferative diabetic retinopathy (PDR) is a late-stage complication of diabetes, responsible for vision loss in diabetic patients. The formation of fibrovascular membranes and scar tissue in the pre-retinal space leads to retinal traction and detachment [1]. Understanding the molecular features of the cells contributing to fibrovascular membrane formation and the cell–cell interactions is crucial for identifying new therapeutic targets that improve treatment approaches and patients’ quality of life.
scRNA-Seq is a powerful technique that allows us to study the molecular profile of each single cell. Since the first paper on scRNA-Seq was published in 2009, studies on this technique have provided insightful information in several fields, followed by exponential growth in the last decade [2–8]. In the context of retinal biology, this approach was fundamental for building a comprehensive transcriptome atlas of fetal and adult retinas and for dissecting the retinal developmental stages [9–11]. scRNA-Seq assays have also revealed the complexity and heterogeneity of the different cell types in the retina and aided in uncovering cell-specific gene expression changes in pathological conditions [12–17]. Single-cell transcriptomics also enables the study of individual pathogenic cell populations in the context of fibrosis at quite high resolution [18–22].
Despite significant improvements, scRNA-Seq is still a challenging technique [23]. Generating high-quality single-cell suspensions from any tissue is critical to preserve their expression profile and ensure meaningful downstream transcriptome data analysis. Several parameters in the dissociation protocol can compromise the viability of the cells and potentially impact the quality of the scRNA-Seq data. Preparing cell suspensions might be difficult for tissue samples, especially for fibrotic tissue, where gentle dissociation might not be sufficient to break the fibers of scar tissue, leading to inefficient cell yield after processing. On the other hand, an excessively harsh digestion might differentially damage cells that are more fragile [23,24].
Papain-based protocols have been shown to successfully dissociate retinal tissue and have been described in several scRNA-Seq studies of retina tissue and organoids [16,23,25]. Another enzyme described for tissue digestion is Collagenase (I and IV), which was described to digest fibrovascular membranes from proliferative diabetic retinopathy patients as well as mouse retinas and other tissues into single-cell suspensions [18,21,26–28]. In our experiments, we used a Collagenase I–based protocol [29].
Another important factor in the scRNA-Seq experiments is the downstream data analyses. Throughout the analysis, it is important to include filtering steps in data analysis processing with the aim of excluding low-quality cells or empty droplets that have very few genes and cell doublets that may exhibit an aberrantly high gene count.
Our protocol is quite efficient in digesting the fibrovascular membranes from PDR patients [29], and we were able to preserve a lot of cell types with a high number of genes detected per cell.
Materials and reagents
Biological materials
Fibrovascular membranes surgically removed from proliferative diabetic retinopathy (PDR) patients
Reagents
Collagenase Type I (Worthington, catalog number: LS004194)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906)
Ethylenediaminetetraacetic acid (EDTA) (Amresco, catalog number: E522)
Illumina TruSeq stranded mRNA (Illumina, catalog number: 20020594)
Chromium Single Cell 3' v3 kit (10X Genomics, catalog number: PN-1000268)
Hank's balanced salt solution (HBSS) (Thermo Fisher Scientific, catalog number: 14025092)
1× Dulbecco’s phosphate-buffered saline (DPBS) with calcium and magnesium (Thermo Fisher Scientific, catalog number: 14040133)
1× Dulbecco’s phosphate-buffered saline (DPBS) without calcium and magnesium (Thermo Fisher Scientific, catalog number: 14190094)
Solutions
Collagenase I solution (see Recipes)
Bovine serum albumin (BSA)-EDTA (see Recipes)
Recipes
Collagenase I solution
Reagent Final concentration Quantity or Volume Collagenase type I 2 mg/mL 500 µL/sample PBS with Ca2+ and Mg2+ Bovine serum albumin (BSA)-EDTA
Reagent Final concentration Quantity or Volume Bovine serum albumin (BSA) 1% 500 µL/sample EDTA 2 mM PBS w/o Ca2+ and Mg2+
Laboratory supplies
1.5 mL microcentrifuge tubes (Eppendorf, catalog number: 022431021)
1 mL pipette (USA Scientific, catalog number: 7110-1000)
Laboratory tips (TipOne, catalog number: 1111-2720)
Equipment
Agilent 2100 Bio-analyzer
Illumina HiSEQ4000
Microcentrifuge (Eppendorf, model: 5424R)
Software and datasets
R software (free download from https://cran.r-project.org/bin/windows/base/) (R version 4.3.2, 2023-10-31)
RStudio (free download from https://posit.co/download/rstudio-desktop/) (Version 2023.03.1+446 (2023.03.1+446)
Seurat (version 5.1.0) and Monocle (Version 2.32.0) packages for RStudio
All raw data have been deposited to GEO (accession no. GSE245561) and trajectory inference analysis code was deposited on GitHub (https://github.com/katiacoranoscheri/PDR-trajectory-analysis).
Procedure
文章信息
稿件历史记录
提交日期: Jun 5, 2024
接收日期: Sep 5, 2024
在线发布日期: Sep 29, 2024
出版日期: Oct 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Scheri, K. C., Tedeschi, T. and Fawzi, A. A. (2024). Single Cell Isolation from Human Diabetic Fibrovascular Membranes for Single-Cell RNA Sequencing. Bio-protocol 14(20): e5096. DOI: 10.21769/BioProtoc.5096.
分类
细胞生物学 > 单细胞分析
生物信息学与计算生物学
分子生物学 > RNA > RNA 测序
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link