- EN - English
- CN - 中文
Host Receptor Pili for Cryo-EM Single-Particle Reconstruction
宿主受体菌毛用于冷冻电镜单颗粒重构
发布: 2024年10月20日第14卷第20期 DOI: 10.21769/BioProtoc.5094 浏览次数: 1586
评审: Alba BlesaCuncai GuoSrajan Kapoor

相关实验方案

通过离子交换色谱、凝胶过滤和MDPF-明胶-酶谱分析法准备和纯化β-1,3-葡聚糖连接的光滑念珠菌细胞壁蛋白酶
Pirjo Pärnänen [...] Pirjo Nikula-Ijäs
2024年03月20日 1706 阅读
通过制备连续聚丙烯酰胺凝胶电泳和凝胶酶谱分析法纯化来自梭状龋齿螺旋体的天然Dentilisin复合物及其功能分析
Pachiyappan Kamarajan [...] Yvonne L. Kapila
2024年04月05日 2080 阅读
Abstract
Single-stranded RNA bacteriophages (ssRNA phages) infect their hosts by binding to the host receptor pili. Purification of pili usually involves mechanical shearing of pili from cells followed by precipitation. However, previous methods often result in low efficiency or unstable results due to pili retraction. This protocol presents an optimized method for purifying receptor type IV pili from Acinetobacter genomospecies 16 (A. gp16), incorporating enhancements in shearing and collection steps to achieve high yields. We found that repeated passage through syringe needles increases yield, and temperature control is crucial during purification. Additionally, the CsCl density gradient was optimized specifically for this specific strain. The purified type IV pili are suitable for cryogenic electron microscopy (cryo-EM) and various biochemical experiments.
Key features
• Pili purification for single-particle cryo-electron microscopy (Cryo-EM) analysis
• This protocol builds upon the F-pili purification method developed by Costa et al. [1] extending its application to the Acinetobacter genomosp. 16.
• It is optimized for higher and more stable pili yields, as well as increased reproducibility.
• The method is tested on various bacterial species and can be adapted to purify different types of pili.
Keywords: Host receptor (宿主受体)Graphical overview
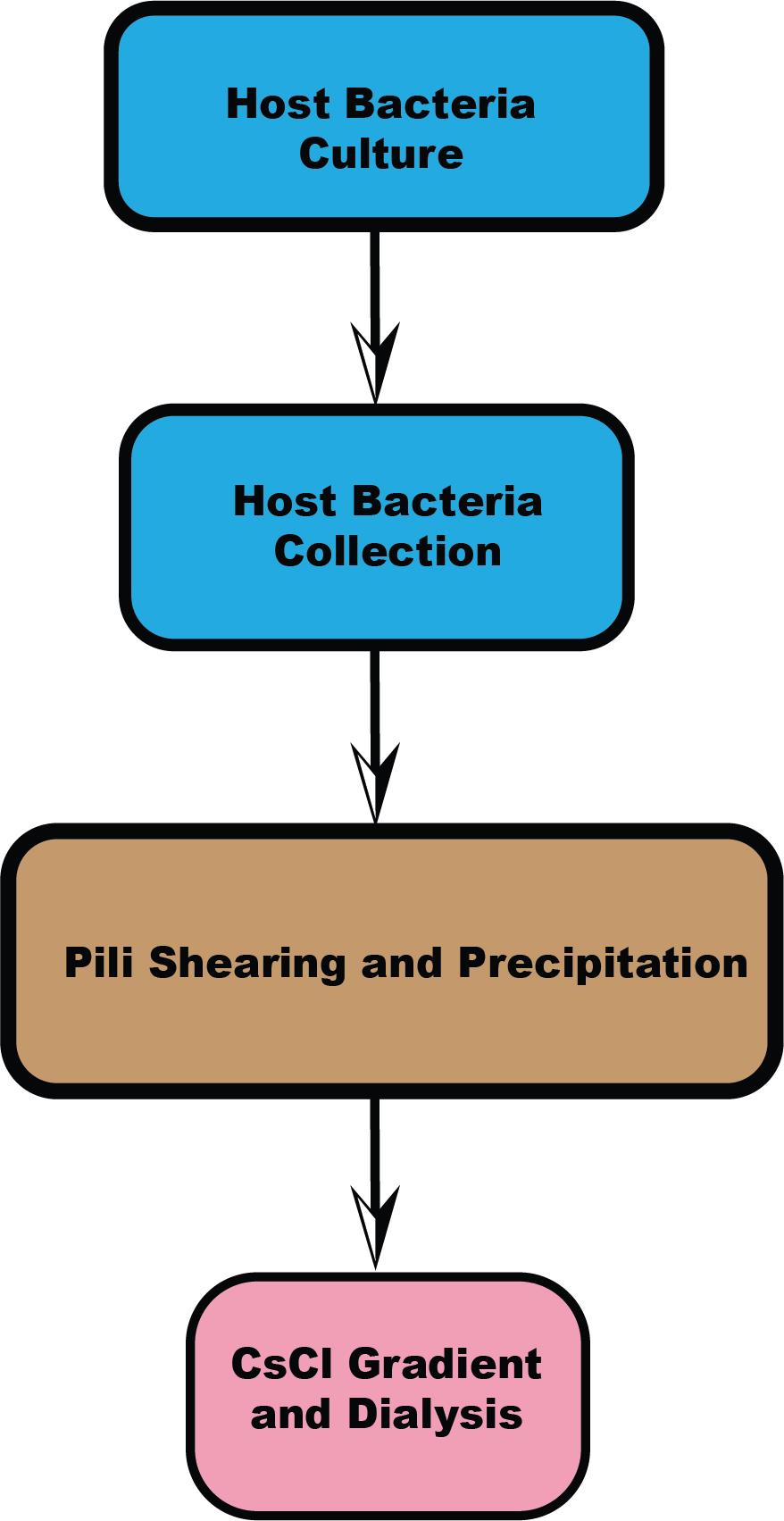
Workflow overview
Background
Acinetobacter, a Gram-negative bacterium, is an aerobic, non-flagellated, and widely distributed coccobacillus [2]. Acinetobacter baumannii, Acinetobacter calcoaceticus, Acinetobacter pittii, and Acinetobacter nosocomialis are opportunistic pathogens known for causing hospital-acquired infections. Bacteria can invade tissue and evade immune response through various mechanisms, including the production of enzymes. Adherence to the host is facilitated by structures such as pili [3–5]. Each year, U.S. hospitals report around 12,000 infections caused by multidrug-resistant Acinetobacter species. The protein secretion processes in Acinetobacter, which are critical targets for vaccine development, are not well understood. Known protein secretion systems in Acinetobacter include the Type II secretion system (T2SS), Type VI secretion system (T6SS), Type IV pilus secretion system, autotransporters, and outer membrane vesicles (OMVs) [6].
In this protocol, we focus on the type IV pili purification from Acinetobacter genomosp. 16 (A. gp16, NCBI: txid70347), which is a target host receptor of single-stranded RNA phage AP205 (ssRNA phage AP205, a virus that can infect Acinetobacter) [7]. Given the crucial role retractile pili play in the virulence of many bacteria, if infection-dependent pili detachment is a common feature of ssRNA phages, these simple viruses could potentially be used to inhibit retractile pili deployment and thereby decrease the virulence of numerous significant bacterial infections [7]. The purified pili can not only be utilized for Cryo-EM structural studies but also for biochemical or biophysical experiments to test protein binding affinity.
Type IV pili are extracellular helical appendages, typically with a diameter of 6–9 nm [8], which are composed of thousands of noncovalently linked pilin subunits. The major pilin of Acinetobacter type IV pili is the PilA monomer, which is composed of an N-termini hydrophobic α-helix, an α-β loop for connection, and the C-termini soluble β-sheet. The mature pilins assemble into type IV pilus by type IV pilus secretion system. Pilins bury the N-terminus helix inside, forming the pilus’s central axis, with the C-terminus headgroup (β-sheet) exposed outside [9].
Several protocols have been established for the purification of different types of pili [1,10,11]. While these methods share commonalities, specific steps can vary depending on the bacterial strain. We tested various pili purification techniques and found that the method developed by Costa et al. [1] for Escherichia coli (E. coli) F-pili provided higher efficiency and yield than other methods. First, we optimized and highlighted critical steps within this protocol to improve and stabilize yield, such as using a syringe to facilitate pili detachment from cells and controlling temperature to prevent pili retraction. Second, we focused on optimizing the procedure for Acinetobacter pili purification, allowing for the collection and separation of two different types of pili simultaneously. There are two types of pili outside the cell boundary: type IV pili and FilA filament. Lastly, this paper outlines a four-section process and provides two options (one for hyper-piliated strains, the other for low-yield strains), making it suitable for pili from other bacteria species as well (Graphical overview). We found that some steps could be simplified when bacterial pili yield is high. For instance, Acinetobacter pili have a relatively higher yield compared to F-pili, which have only one or two pili per cell. Microscope negative staining can be used to visually assess the number of pili per cell, determining whether the strain is hyper-piliated or has a lower yield. Based on this assessment, one can decide whether to follow the method in Sections B and C or Sections B2 and C2. Sections B and C provide a quicker method for obtaining pili from Acinetobacter and other high-yield species. Additionally, alternative steps in Sections B2 and C2, which mainly optimize the method from Costa et al. [1] are provided for strains with lower pili yields (e.g., E. coli F-pili).
In summary, this protocol has been optimized based on previous pili purification methods to enhance yield and stability, particularly for Acinetobacter pili. This method can also be generalized for the purification of pili in other species.
Materials and reagents
Biological materials
Acinetobacter genomosp. 16 (A. gp16, NCBI: txid70347)
Reagents
Difco LB broth, Miller (Miller Broth) 500 g (BD Bioscience, catalog number: 244620)
Tris (Bio-Rad, catalog number: 1610719)
Sodium chloride (NaCl) (crystalline/certified ACS) (Fisher Scientific, catalog number: 7647-14-5)
Sodium citrate dihydrate (granular/certified) (Fisher Scientific, catalog number: 6132-04-3)
(Optional) Polyethylene glycol 6000 (PEG6000) (Thermo Fisher Scientific, catalog number: 25322-68-3)
Cesium chloride (Thermo Fisher Scientific, catalog number: 7647-17-8)
Solutions
SSC buffer (see Recipes)
Dialysis buffer (see Recipes)
5% PEG6000 (see Recipes)
CsCl step gradient buffer (see Recipes)
Recipes
Note: All in sterile water.
SSC buffer
Reagent (room temperature) Final concentration (pH = 7.2) Sodium citrate 15 mM NaCl 150 mM The buffer requires autoclaving.
Dialysis buffer
Reagent (room temperature) Final concentration (pH = 8.0) Tris-HCl 50 mM NaCl 150 mM The buffer requires autoclaving.
5% PEG6000
Dissolve PEG6000 into dialysis buffer at pH = 8.0. The final volume should be 1 L of dialysis buffer with 50 g of PEG6000. The buffer requires autoclaving.
CsCl step gradient buffer
Reagent (room temperature) Final concentration (pH = 8.0) CsCl The buffer needs different densities to help form the gradient in the centrifuge tube.
Preparing density at 1.0 g/cm3, 1.1 g/cm3, 1.2 g/cm3, 1.3g/cm3, 1.4 g/cm3Dialysis buffer
Laboratory supplies
Slide-A-LyzerTM dialysis cassettes, 20 K MWCO (Thermo Fisher, catalog number: 66003)
Sterile syringes for single use, 5 mL and 3 mL (Fisher Scientific, catalog numbers: 14-955-457, 14-955-458)
Disposable hypodermic needles 25 G, 23 G, and 18 G (Fisher Scientific, catalog numbers: 26406, 26408, 26420)
BD General Use and PrecisionGlide hypodermic needles (Fisher Scientific, catalog number: 305127)
FisherbrandTM Petri dishes with clear lids (Fisher Scientific, catalog number: 0875713)
FalconTM round-bottom polypropylene test tubes with cap (Fisher Scientific, catalog number: 14-959-11B)
PierceTM Protein Concentrators PES, 30 K MWCO (different sizes) (Thermo Fisher Scientific, catalog number: 88502, 88522, 88531)
Equipment
SW 41 Ti swinging-bucket rotor (Beckman, catalog number: 331362)
13.2 mL, open-top thin-wall ultra-clear tube, 14 × 89 mm, 50 Pk (Beckman, catalog number: 344059)
Procedure
文章信息
稿件历史记录
提交日期: Jun 22, 2024
接收日期: Aug 27, 2024
在线发布日期: Sep 25, 2024
出版日期: Oct 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Meng, R. (2024). Host Receptor Pili for Cryo-EM Single-Particle Reconstruction. Bio-protocol 14(20): e5094. DOI: 10.21769/BioProtoc.5094.
分类
微生物学 > 微生物生物化学 > 蛋白质 > 分离和纯化
生物物理学 > 电子冷冻断层扫描
生物化学 > 蛋白质 > 自组装
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link