- EN - English
- CN - 中文
Genetic Tagging and Imaging of Proteins with iFAST in Candida albicans
使用iFAST在白色念珠菌中进行蛋白质的基因标记与成像
发布: 2024年10月05日第14卷第19期 DOI: 10.21769/BioProtoc.5082 浏览次数: 1484
评审: Shailesh KumarLucy XieAnonymous reviewer(s)
Abstract
Candida albicans is the most common human fungal pathogen, able to reside in a broad range of niches within the human body. Even though C. albicans systemic infection is associated with high mortality, the fungus has historically received relatively little attention, resulting in a lack of optimized molecular and fluorescent tools. Over the last decade, some extra focus has been put on the optimization of fluorescent proteins (FPs) of C. albicans. However, as the FPs are GFP-type, they require an aerobic environment and a relatively long period to fully mature. Recently, we have shown the application of a novel type of fluorogen-based FP, with an improved version of fluorescence activating and absorption shifting tag (iFAST), in C. albicans. Due to the dynamic relation between iFAST and its fluorogens, the system has the advantage of being reversible in terms of fluorescence. Furthermore, the combination of iFAST with different fluorogens results in different spectral and cellular properties, allowing customization of the system.
Key features
• Genetic integration and tagging with the iFAST tag in Candida albicans.
• Imaging and localization of a protein of interest tagged with iFAST.
• Reversibility of fluorescence with iFAST.
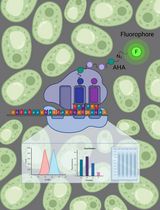
Keywords: Candida albicans (白色念珠菌)Graphical overview
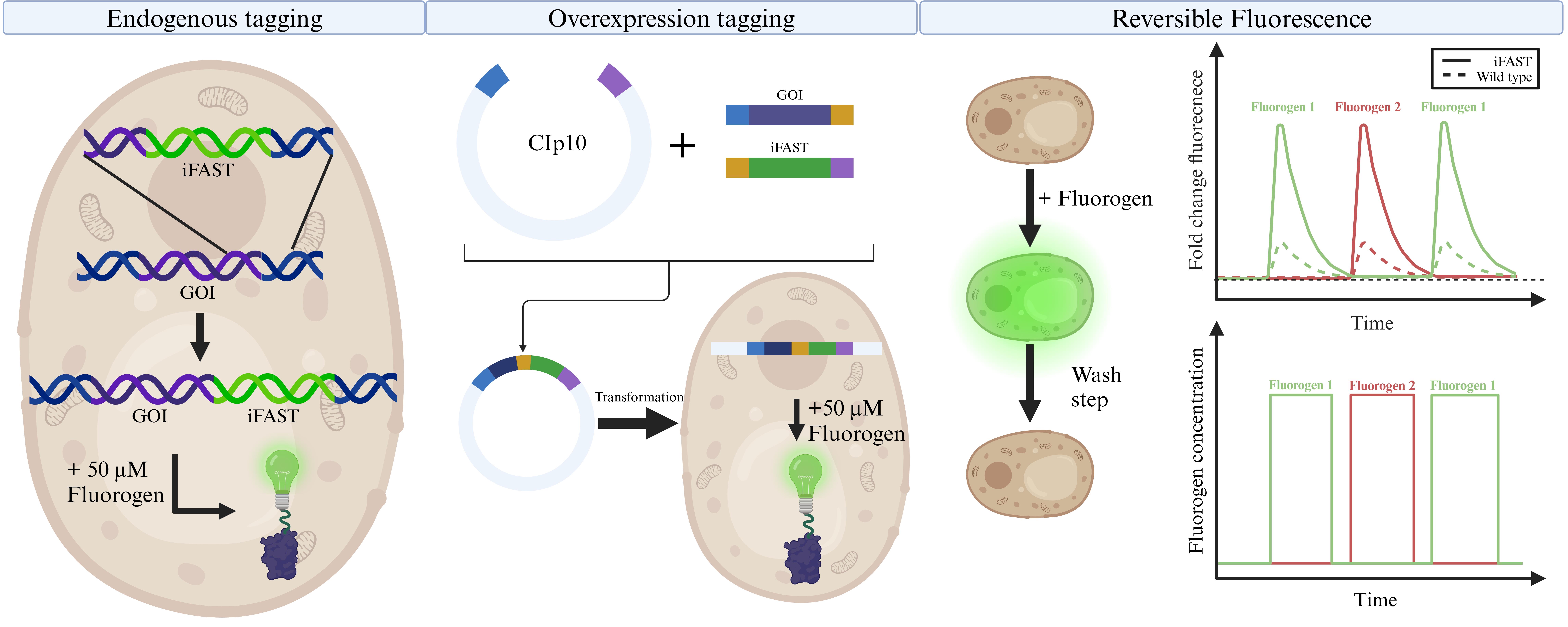
Background
The ubiquitous commensal Candida albicans is a fungus that resides in the gastrointestinal tract, blood, skin, and other mucosal surfaces [1–3]. In healthy individuals, the immune system is sufficiently capable of eradicating C. albicans, but upon opportunistic circumstances, the fungus can become pathogenic, often with a high morbidity rate [4]. As a species, C. albicans is closely related to the eukaryotic model organism Saccharomyces cerevisiae. However, despite this close relationship, C. albicans has an aberrant codon usage, translating the CUG codon to serine instead of leucine in 96% of cases [5,6]. Consequently, heterologous expression of C. albicans proteins requires codon optimization. Conversely, this also means that molecular tools and protein labeling techniques developed for use in S. cerevisiae cannot easily be translated to C. albicans, as these will suffer from the same mistranslation issues. Regarding this issue, several techniques for codon optimization in C. albicans have been reported [7,8], but studies have shown that these are not generally applicable [9].
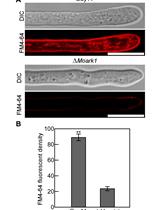
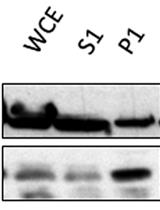
In recent years, some fluorescent tools have been developed/optimized for use in C. albicans (and extensively reviewed in Van Genechten et al. [10]). In this protocol, we show the use of a novel type of fluorescent protein, an improved version of fluorescence activating and absorption shifting tag (iFAST), which requires the binding of a fluorogen, instead of chromophore maturation, to be fluorescently active. This novel system was developed by the group of Gautier for use in mammalian cells [11,12]. The iFAST protein is half the size of GFP and has the added benefit of reversibly binding its fluorogens, which are analogs of 4-hydroxybenzyliden-rhodanine (HBR). On their own, iFAST and HBR analogs are relatively non-fluorescent, but upon contact of iFAST protein with its fluorogen, fluorescence can be observed. An added benefit of this system is that the interaction is reversible and does not require oxygen, making this a reversible fluorescent system even in anaerobic conditions, where classical FPs like GFP do not work.
Materials and reagents
Biological materials
SC5314 Candida albicans wild-type strain [13]
SN152 Candida albicans arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ::imm434 IRO1/iro1Δ::imm434 [14]
Reagents
Granulated yeast extract (Merk, catalog number: 1.03753)
Bacterial peptone (Oxoid, catalog number: LP0037B)
Glucose (Fluka, catalog number: 49459)
Glycerol (Sigma, CAS number: 58-81-5)
Granulated bacteriological agar (Difco, catalog number: 214530)
Nourseothricin (Jena Bioscience, CAS number: 96736-11-7)
Complete synthetic mixture (CSM) (MP Biomedicals, catalog number: 114500022)
Yeast nitrogen base (YNB) without amino acids, ammonium sulfate, and riboflavin (Formedium, catalog number: YN6501)
Ammonium sulfate (VWR, CAS number: 7783-20-2)
Lithium acetate dihydrate (LiAc·2H2O) (Sigma-Aldrich, CAS number: 6108-17-4)
Polyethylene glycol (PEG) 3350 (Sigma-Aldrich, CAS number: 25322-68-3)
Trizma® base (Sigma-Aldrich, CAS number: 77-86-1)
HCl (Sigma-Aldrich, CAS number: 7647-01-0)
EDTA (Sigma-Aldrich, CAS number: 6381-92-6)
HMBR (Twinkle Factory, catalog number: 480541-250)
HBR-3,5DM (Twinkle Factory, catalog number: 499558-250)
HBR-3,5DOM (Twinkle Factory, catalog number: 516600-250)
HIFI Q5 polymerase (New England Biolabs, catalog number: NEB M0491L)
NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, catalog number: NEB E2621X)
ssDNA, MB-grade from fish sperm (Merck, catalog number: 11467140001)
rCutsmart restriction buffer (New England Biolabs, catalog number: B6004S)
Restriction enzymes:
StuI (New England Biolabs, catalog number: R0187S)
NheI-HF (New England Biolabs, catalog number: R3131S)
PstI-HF (New England Biolabs, catalog number: R3140S)
Solutions
YPD (see Recipes)
YPD-NAT agar (see Recipes)
YPD-glycerol (see Recipes)
Low fluorescence medium (see Recipes)
1 M LiAc (see Recipes)
PEG 3350 (50%) (see Recipes)
TE (10×) (see Recipes)
LiAc/TE buffer (see Recipes)
LiAc/PEG buffer (see Recipes)
5 mM fluorogen stock solution (see Recipes)
Recipes
YPD (1 L)
Reagent Final concentration Quantity or Volume Yeast extract granulated 1% (w/v) 10 g Bacteriological peptone 2% (w/v) 20 g Glucose 2% (w/v) 20 g H2O (demineralized) n/a 1 L YPD-NAT agar (1 L)
*Note: Add the nourseothricin after heat sterilization and when the medium has cooled down to approximately 50 °C.
Reagent Final concentration Quantity or Volume Yeast extract granulated 1% (w/v) 10 g Bacteriological peptone 2% (w/v) 20 g Glucose 2% (w/v) 20 g Bacteriological agar 2% (w/v) 20 g Nourseothricin* 0.02% (w/v) 200 mg H2O (demineralized) n/a 1 L YPD-glycerol (100 mL)
Reagent Final concentration Quantity or Volume Yeast extract granulated 1% (w/v) 1 g Bacteriological peptone 2% (w/v) 2 g Glycerol 30% (v/v) 30 mL H2O (demineralized) n/a 70 mL Low fluorescence medium (1 L)
Reagent Final concentration Quantity or Volume CSM 0.079% (w/v) 0.79 g YNB (without amino acids, without ammonium sulfate, without riboflavin) 0.69% (w/v) 6.9 g Ammonium sulfate 0.5% (v/v) 5 g Glucose 2% (w/v) 20 g H2O (demineralized) n/a 1 L 1 M LiAc (100 mL)
Reagent Final concentration Quantity or Volume LiAc·2H2O 1 M 10.2 g H2O (demineralized) n/a 100 mL PEG 3550 (50%) (100 mL)
*Note: First, add 50 mL of H2O and a stir bar. Let the PEG dissolve overnight and add water up to a total volume of 100 mL.
Reagent Final concentration Quantity or Volume PEG 3550 50% (w/v) 50 g H2O (demineralized) n/a Until 100 mL* TE (10×) (1 L)
*Note: After adding Tris, adjust the pH to 8 using HCl.
Reagent Final concentration Quantity or Volume Tris 10 mM 12.11 g* EDTA 1 mM 2.92 g H2O (demineralized) n/a 1 L LiAc/TE buffer (1 mL)
Reagent Final concentration Quantity or Volume LiAc (1M) 0.1 M 100 µL TE (10×) 1× 100 µL H2O (MiliQ) n/a 800 µL LiAc/PEG buffer (1 mL)
Reagent Final concentration Quantity or Volume LiAc (1 M) 0.1 M 100 µL TE (10×) 1× 100 µL PEG 3350 [50% (w/v)] 40% (w/v) 800 µL Fluorogen stock solution
*Note: The quantity is calculated to make a total of 100 mL. Store this stock solution in aliquots of 20 µL at -20 °C. Make a separate stock solution for each fluorogen; do not mix. Fluorogen stock solutions can withstand multiple freeze-thaw cycles; however, try not to defrost more than five times. Additionally, it is best to protect the fluorogens from light as much as possible when working.
Reagent Final concentration Quantity or Volume HMBR 5 mM 0.125 g* HBR-3,5DM 5 mM 0.133 g* HBR-3,5DOM 5 mM 0.149 g*
Laboratory supplies
Microtubes (1.5 mL) (Eppendorf, catalog number: 0030125150)
Micro pipettes (sizes 1,000, 200, and 10 µL)
Micro pipette tips (standard tips of 1,000, 200, and 10 µL)
Microscope slides (VWR, catalog number: 630-1985)
Glass covers (VWR, catalog number: 631-0122)
Glass culture tubes (10 mL) (Fisher Scientific, catalog number: 11557403)
Caps for culture tubes (VWR, catalog number: LUDI184010631)
Erlenmeyer flasks (300 mL) (VWR, catalog number: 391-0275)
Erlenmeyer flask caps (VWR, catalog number: 391-0950)
Clear cuvette (VWR, catalog number: 97000-590)
Vortex (VWR, catalog number: 444-1372)
NucleoSpinTM Gel and PCR Clean-up Kit (Macherey-Nagel, catalog number: 740611.50)
NucleoSpinTM Plasmid EasyPure Kit (Macherey-Nagel, catalog number: 740727.50)
Equipment
FV1000 microscope (Olympus, model: FV1000-IX81) with 60× UPlanSApo (NA1.35) objective lens (Olympus) or equivalent
ThermoMixer® F1.5 (Eppendorf, catalog number: 5384000020) or equivalent
Shaking incubator (New Brunswick Scientific, model: Innova40) or equivalent
Microtube centrifuge (Eppendorf, model: 5471C) or equivalent
BioPhotometer (Eppendorf, model: 6131)
NanoDropTM 1000 spectrophotometer (Thermo Scientific, model: ND-1000) or equivalent
Software and datasets
FV10-ASW 4.2 software package (Olympus)
ImageJ (v1.53t, 24/08/2022) (https://imagej.net/software/fiji/downloads)
RStudio (R version: 4.3.2, 31/10/2023) (https://cran.rstudio.com/; https://posit.co/download/rstudio-desktop/)
Procedure
文章信息
稿件历史记录
提交日期: May 30, 2024
接收日期: Aug 23, 2024
在线发布日期: Sep 10, 2024
出版日期: Oct 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Devos, J., Van Dijck, P. and Van Genechten, W. (2024). Genetic Tagging and Imaging of Proteins with iFAST in Candida albicans. Bio-protocol 14(19): e5082. DOI: 10.21769/BioProtoc.5082.
分类
微生物学 > 微生物细胞生物学 > 细胞染色
分子生物学 > 蛋白质 > 检测
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link