- EN - English
- CN - 中文
FixNCut: A Practical Guide to Sample Preservation by Reversible Fixation for Single Cell Assays
FixNCut:单细胞检测样本可逆固定保存实用指南
(§ Technical contact) 发布: 2024年09月05日第14卷第17期 DOI: 10.21769/BioProtoc.5063 浏览次数: 3284
评审: Pilar Villacampa AlcubierreLeonor GouveiaSaba Asam
Abstract
The quality of standard single-cell experiments often depends on the immediate processing of cells or tissues post-harvest to preserve fragile and vulnerable cell populations, unless the samples are adequately fixed and stored. Despite the recent rise in popularity of probe-based and aldehyde-fixed RNA assays, these methods face limitations in species and target availability and are not suitable for immunoprofiling or assessing chromatin accessibility. Recently, a reversible fixation strategy known as FixNCut has been successfully deployed to separate sampling from downstream applications in a reproducible and robust manner, avoiding stress or necrosis-related artifacts. In this article, we present an optimized and robust practical guide to the FixNCut protocol to aid the end-to-end adaptation of this versatile method. This protocol not only decouples tissue or cell harvesting from single-cell assays but also enables a flexible and decentralized workflow that unlocks the potential for single-cell analysis as well as unconventional study designs that were previously considered unfeasible.
Key features
• Reversible fixation: Preserves cellular and molecular structures with the option to later reverse the fixation for downstream applications, maintaining cell integrity
• Compatibility with single-cell assays: Supports single-cell genomic assays such as scRNA-seq and ATAC-seq, essential for high-resolution analysis of cell function and gene expression
• Flexibility in sample handling: Allows immediate fixation post-collection, decoupling sample processing from analysis, beneficial in settings where immediate processing is impractical
• Preservation of RNA and DNA integrity: Effectively preserves RNA and DNA, reducing degradation to ensure accurate transcriptomic and genomic profiling
• Suitability for various biological samples: Applicable to a wide range of biological samples, including tissues and cell suspensions, whether freshly isolated or post-dissociated
• Enables multi-center studies: Facilitates collaborative research across multiple centers by allowing sample fixation at the point of collection, enhancing research scale and diversity
• Avoidance of artifacts: Minimizes stress or necrosis-related artifacts, preserving the natural cellular physiology for accurate genomic and transcriptomic analysis
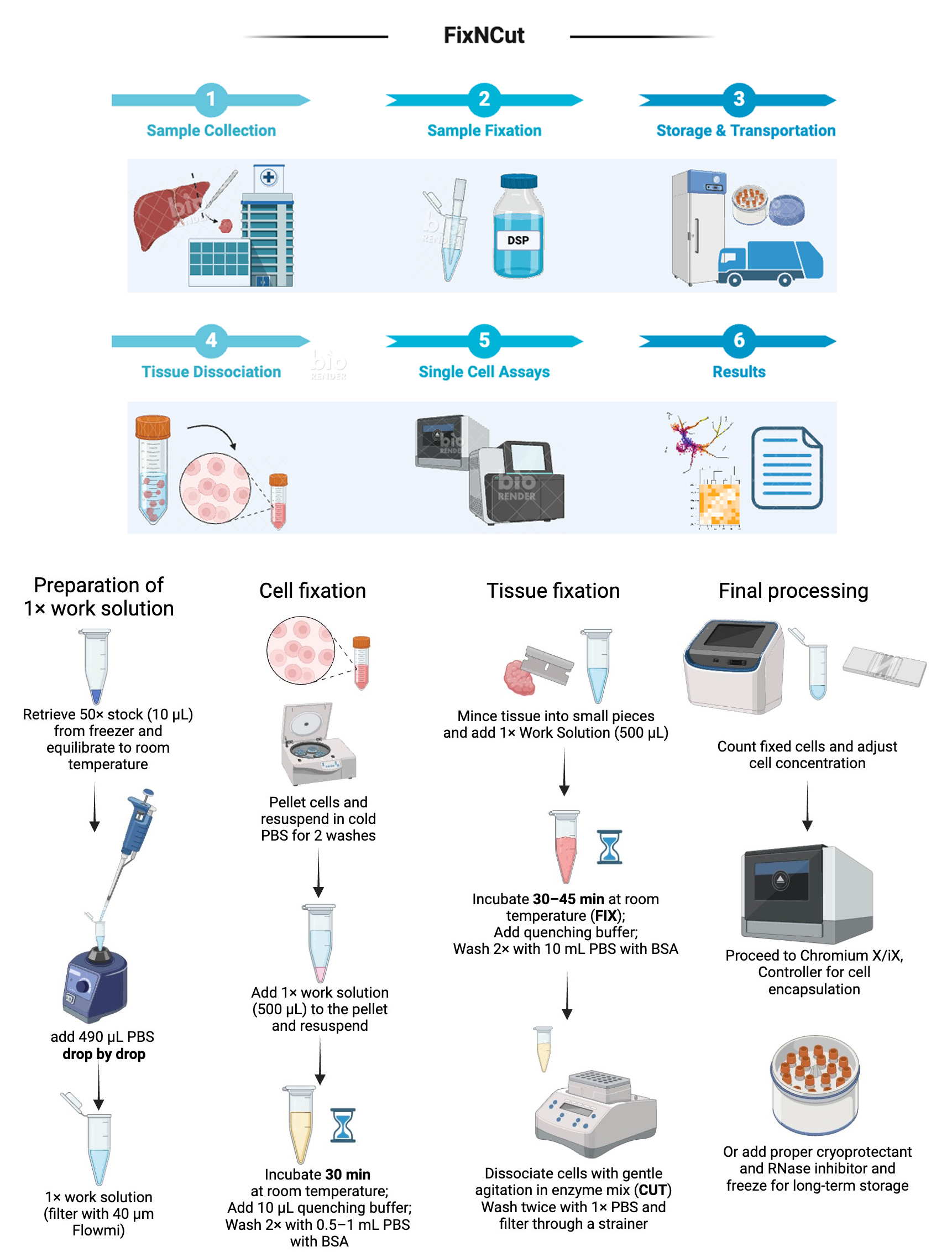
Keywords: Single cell (单细胞)Graphical overview
Background
Single-cell techniques have revolutionized biological and clinical research by quantitatively capturing the genomic and transcriptomic state of individual cells at unprecedented resolution and scope [1]. This approach moves beyond averaging the bulk population, which consists of various cell types. However, the technical and logistical challenges of acquiring or generating high-viability single-cell suspensions have limited the application of single-cell genomics and transcriptomics.
Multi-center clinical studies often involve sample collection from remote areas lacking processing infrastructure [2,3]; additionally, longitudinal and cohort recruitment of patients occurs over the years, subject to patient availability and logistics constraints. Moreover, even in advanced clinical centers, the urgency of tissue harvesting during surgical procedures, biopsies, or autopsies can depend on factors such as the type of surgery, the purpose, and the specific protocol followed, as well as the transfer procedures between surgery and histopathology [4,5]. In basic research using organoids, embryos, or rare specimens, the limited cell number poses a significant bottleneck for conducting scientifically meaningful and economically feasible single-cell analysis.
In many experimental setups typical for clinical research, the freeze-thaw process remains the predominant option for sample collection en masse. However, freezing will physically damage the cell membrane, impact the post-thaw viability, and disrupt intracellular compartments where endogenous RNases are sequestered, leading to RNA degradation. In addition, cryopreservation may lead to the loss of certain cell types and induce cellular stress response [6]. Moreover, variably staggered freezing and thawing through aliquoting cells for parallel orthogonal assays, for example, single-cell mass cytometry (CyTOF), single-cell RNA-Seq, single-cell ATAC-Seq, and additional multiomic assays, can lead to inconsistency, introducing technical artifacts and batch-wise noise to the data. Recently, the fixed RNA (flex) assay has been gaining popularity due to its robustness; however, so far probes are only available for humans and mice and do not target immunologically and clinically paramount transcripts such as joining and variable regions in B and T-cell receptor (BCR and TCR), Killer cell immunoglobulin-like receptors (KIRs), and human leukocyte antigen (HLA), nor polyadenylated non-coding RNA.
Notably, many important immune cells—such as neutrophils, dendritic cells, monocytes, macrophages, and lymphocytes (B cell and T cells), as well as cells with higher intrinsic mechanical integrity such as epithelial cells—are considered fragile for single-cell applications due to their susceptibility to mechanical damage during sample preparation, including cell sorting or tissue dissociation [7–9]. Loss of these cells often leads to underrepresentation in downstream analysis. Therefore, identifying a method allowing the stabilization of fragile cells without perturbing RNA integrity will enhance the recovery of these important populations.
The ideal fixative must possess several characteristics: first, it should be small and able to penetrate the cell membrane and tissues. Second, it should preserve the structure and integrity of cells. Third, it should not destroy RNA integrity and should, ideally, inhibit RNase activity at least temporarily. Currently, one of the most common options for single-cell assays is using either standard or modified methanol fixation [10–14]. Methanol works by dehydrating samples and denaturing proteins in a gentle manner, similar to histology fixation. However, methanol as a standalone fixative has raised concerns regarding biased results or ambient RNA leakage in single-cell RNA-Seq [6]. A possible mechanistic explanation is the irreversible intracellular compartment disruption resulting in the loss of normal lipid and protein structure during dehydration–rehydration cycles. In addition, incomplete reverse transcription of mRNAs with more complex secondary structures was suggested as a major caveat [15].
In contrast, dithio-bis-succinimidyl propionate (DSP), known as Lomant's reagent [16], has been traditionally used for histological tissue fixation to stabilize cellular integrity and structures through covalent crosslinking of free amine groups found at the N-terminus of polypeptide chains (diagrammatic molecular basis is depicted in Figure 1 of Akaki et al. [17]). It has been repurposed for single-cell RNA sequencing by maintaining RNA integrity and yield in bulk RNA extractions [18–20]. The cell and membrane permeability have been proven to be particularly useful in detecting rare cell populations such as tissue-resident immune cells. Certain T-cell populations can exist in very low numbers upon activation, making them difficult to isolate and analyze with conventional single-cell methods. A protocol called CLInt-Seq (crosslinker regulated intracellular phenotype sequencing) was developed to overcome the hurdle by crosslinking intracellular cytokines [21]. This technique combines and improves upon existing techniques to collect and genetically sequence rare T cells. The reversibility of DSP-induced crosslinking is a key feature allowing cells and tissue to be further processed or dissociated for downstream applications. However, a recent study concluded that de-crosslinking the DSP-fixed samples is optional, and additional handling steps of de-crosslinking may contribute counterproductively to cell loss [22].
Recently, we developed the further optimized FixNCut protocol and demonstrated its robustness and benefits systematically by comparing fresh and fixed lung, colon, and pancreas samples from different species even under cryopreservation for an extended period [23,24]. Here, we provide an end-to-end solution in the form of a practical guide on the implementation of this method, to remove practical barriers by streamlining the transport of samples and scheduling of shared instruments for downstream single-cell isolation and processing.
Materials and reagents
Biological materials
Tissue samples and cell suspensions can both be used. For cells, the protocol is set up for up to 2 × 106 cells. For more cells, scale up the fixation accordingly. Tissues larger than 3 mm in diameter or edge length need to be partitioned into smaller pieces to facilitate fixative penetration
DSP (Dithio-bis-succinimidyl propionate), also known as Lomant's reagent (Thermo Fisher, catalog number: 22586)
DMSO, anhydrous (Thermo Fisher, Molecular Probes, catalog number: D12345)
LiberaseTM research grade, 10 mg (Roche, catalog number: 5401119001)
Protector RNase inhibitor (40 U/μL) (Roche, catalog number: RNAINH-RO)
UltraPureTM 1 M Tris-HCI buffer, pH 7.5 (Thermo Fisher, Invitrogen, catalog number: 15567027)
10× PBS buffer, pH 7.4 (Invitrogen, catalog number: AM9624 or AM9625)
Ambion nuclease-free water (Invitrogen, catalog number: AM9932)
Bovine serum albumin (BSA) solution, sterile filtered and cell-culture tested (Sigma Aldrich, catalog number: A1595)
Tris base, UltraPure Tris buffer (powder format) (Thermo Fisher, Invitrogen, catalog number: 15504020)
Fixation concentrate stock solution (1 mL, for 100 standard assays) (see Recipes)
PBS with 1% BSA (see Recipes)
1 M Tris pH 7.5 (optional, if not using ready-made buffer) (see Recipes)
Fixation concentrate stock solution (1 mL, for 100 standard assays)
Reagent Final concentration Amount DSP (powder) n/a 50 mg DMSO n/a 1 mL Total 50 mg/mL (w/v) 1 mL PBS with 1% BSA
Reagent Final concentration Amount 10× PBS n/a 50 mg BSA 10% n/a 1 mL Total n/a 1 mL 1 M Tris pH 7.5 (optional, if not using ready-made buffer)
Start with 30 mL of water and adjust pH to 7.5 by adding 5 M HCl to a final volume of 50 mL.
Reagent Final concentration Amount Tris base n/a 6.05 g Nuclease-free water n/a 50 mL Total 1 M 50 mL
Laboratory supplies
DNA LoBind tubes 1.5 mL (Eppendorf, catalog number: 022431021)
DNA LoBind tubes 2.0 mL (Eppendorf, catalog number: 022431048)
Flowmi® cell strainers, porosity 40 μm, for 1000 μL pipette tips (Sigma, Scienceware, catalog number: BAH136800040)
pluriStrainer mini 70 μm (pluriSelect, catalog number: 43-10070-40)
Falcon conical centrifuge tubes (Corning, catalog number: 352070 for 50 mL, 352095 for 15 mL)
Sterile serological pipettes with pipettor
Rainin Pipet-Lite LTS pipette tips (Rainin, catalog number: 30389240, 30389213, 30389226)
Equipment
Centrifuge 5810/5810R (Eppendorf, catalog number: EP022628188) with rotor S-4-104
Rotor F-35-6-30 (Eppendorf, catalog number: EP5427716009)
S-4-104 rotor adapters for 50 × 1.5/2 mL tubes (Eppendorf, catalog number: 58-257-40009)
Standard heavy-duty vortex mixer (VWR or Fisherbrand, catalog number: 97043-562)
ThermoMixer C (Eppendorf, catalog number: 05-412-503) or thermoblock with minimal temperature fluctuation
Ice bucket with cool blocks or Cool Rack CFT30 (Corning, catalog number: CLS432052)
Automated cell counter, e.g., Luna-FX7 (Logos Biosystems) or hemocytometer
CoolCell freezing container for 12 × 1 mL or 2 mL cryogenic vials (Corning, catalog number: 432000) or Mr. Frosty freezing container (Thermo Scientific, catalog number: 5100-0001)
Tissue-Tek cold plate (VWR Scientific, catalog number: 25608-942)
Procedure
文章信息
稿件历史记录
提交日期: Jun 1, 2024
接收日期: Jul 28, 2024
在线发布日期: Aug 13, 2024
出版日期: Sep 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Wang, S., Jiménez-Gracia, L., de Amaral, A. A., Vlachos, I. S., Plummer, J., Heyn, H. and Martelotto, L. G. (2024). FixNCut: A Practical Guide to Sample Preservation by Reversible Fixation for Single Cell Assays. Bio-protocol 14(17): e5063. DOI: 10.21769/BioProtoc.5063.
分类
细胞生物学 > 单细胞分析
分子生物学 > DNA > DNA 测序
细胞生物学 > 细胞分离和培养 > 低温贮存
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Can you cryoperserve after DSP fixation and quenching of tissue?
Share
Bluesky
X
Copy link