- EN - English
- CN - 中文
Single-Molecule Sequencing of the C9orf72 Repeat Expansion in Patient iPSCs
患者iPS细胞中C9orf72重复扩展的单分子测序
(*contributed equally to this work) 发布: 2024年09月05日第14卷第17期 DOI: 10.21769/BioProtoc.5060 浏览次数: 2241
评审: Marion HoggSrinidhi Rao Sripathy RaoPrashanth N Suravajhala
Abstract
A hexanucleotide GGGGCC repeat expansion in the C9orf72 gene is the most frequent genetic cause of amyotrophic lateral sclerosis (ALS) and frontal temporal dementia (FTD). C9orf72 repeat expansions are currently identified with long-range PCR or Southern blot for clinical and research purposes, but these methods lack accuracy and sensitivity. The GC-rich and repetitive content of the region cannot be amplified by PCR, which leads traditional sequencing approaches to fail. We turned instead to PacBio single-molecule sequencing to detect and size the C9orf72 repeat expansion without amplification. We isolated high molecular weight genomic DNA from patient-derived iPSCs of varying repeat lengths and then excised the region containing the C9orf72 repeat expansion from naked DNA with a CRISPR/Cas9 system. We added adapters to the cut ends, capturing the target region for sequencing on PacBio’s Sequel, Sequel II, or Sequel IIe. This approach enriches the C9orf72 repeat region without amplification and allows the repeat expansion to be consistently and accurately sized, even for repeats in the thousands.
Key features
• This protocol is adapted from PacBio’s previous “no-amp targeted sequencing utilizing the CRISPR-Cas9 system.”
• Optimized for sizing C9orf72 repeat expansions in patient-derived iPSCs and applicable to DNA from any cell type, blood, or tissue.
• Requires high molecular weight naked DNA.
• Compatible with Sequel I and II but not Revio.
Keywords: C9orf72 (C9orf72)Graphical overview
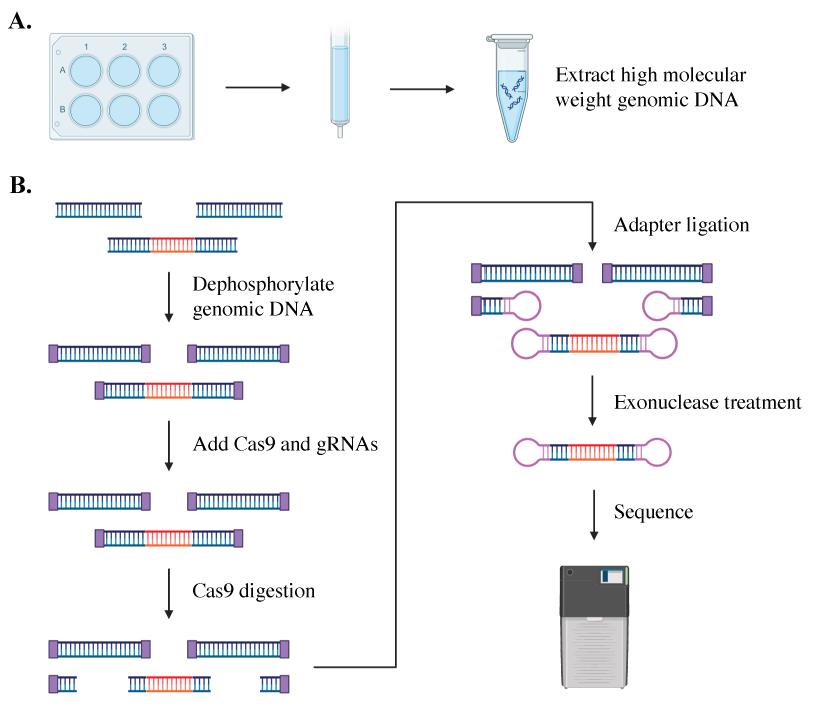
Background
The most frequent genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) is an intronic repeat expansion in the C9orf72 gene [1–3]. While non-diseased alleles typically have 10 or fewer GGGGCC repeats, expanded alleles can have thousands [1,2,4,5]. Accurately measuring the C9orf72 repeat expansion length applies to both clinical diagnosis and answering fundamental questions about the function of C9orf72 in health and disease. Traditional short-read sequencing methods fail to size the C9orf72 repeat region because amplification fails across the GC-rich region of the first intron of C9orf72, and short-read sequencing does not provide unique sequences adjacent to GGGGCC repeats to permit their alignment [5]. Instead, long-range PCR [6] and Southern blot [7] are used as clinical diagnostic and research tools to identify C9orf72 repeat expansions, but they are limited. Long-range PCR fails for repeats greater than 145 and, while Southern blot can identify long repeats, it requires a large amount of input DNA [5].
Single-molecule sequencing is an accurate, reliable, and sensitive method to size repeat lengths of varied sizes, showing success in sequencing the C9orf72 repeat expansion in both plasmid and tissue [5,8–10]. This amplification-free process employs a CRISPR/Cas9 system to excise and sequence the region containing the C9orf72 expansion in high-quality DNA from patient iPSCs (see Graphical overview). The excised genomic fragment is then sequenced from end to end, providing phased sequencing of the targeted region without the need for bioinformatic imputation of its sequence or structure. In addition to identifying and quantifying repeat expansions of various lengths, the sequencing data provides insights into the gene structure. Single-molecule sequencing can identify mosaicism within samples and help determine the stability of repeat lengths over time [5,8,10]. It can be used to assess cell line clonality, editing outcomes, and differences between each allele of a gene in a single individual [5,10]. This method provides additional information on the methylation of each allele within an individual or across a population [10].
Despite the technological advances afforded by single-molecule sequencing, there are a few limitations to employing this method in research and clinical settings. The first is cost. Single-molecule sequencing is expensive. The ability to multiplex samples, as we describe here, reduces the cost of this protocol. The second limitation is the variability in read depth with longer repeat expansions. As the size of repeat expansion increases, the number of reads on the expanded allele tends to decrease [5]. Long expansions can still be sized, albeit with reduced read depth. Over time, as technology improves and the number of reads for each run increases, this bias is likely to decrease.
We have optimized the protocol presented here for genomic DNA extracted from human induced pluripotent stem cell (iPSC) lines derived from C9-ALS/FTD patient samples. This protocol can also be applied to other sources of DNA such as any cultured cell type as well as donor blood and fresh or frozen tissue. This method can also be used to target other repeat expansions such as TCF4 and DPMK, for which this approach was initially reported [11,12], or other genomic regions of interest by targeting the Cas9/gRNA excision to the target region. This protocol is compatible with Sequel I and Sequel II. PacBio’s PureTargetTM Repeat Expansion Panel [13], which is an improved version of the NoAmp protocol, can be used additionally with Revio and quantifies 20 different repeat expansions for targeted single-molecule sequencing simultaneously and in the same individual.
Materials and reagents
Biological materials
5–25 μg of DNA from induced pluripotent stem cells (iPSCs) with C9orf72 repeat expansions (also compatible with DNA from blood or tissue)
Reagents
DPBS (Gibco, catalog number: 14190235)
ReLeSR (STEMCELL Technologies, catalog number: 05872)
Genomic DNA buffer set (QIAGEN, catalog number: 19060)
Proteinase K (Qiagen, catalog number: 19131)
QubitTM 1× dsDNA HS Assay kit (Thermo Fisher Scientific, catalog number: Q33230)
Agarose (Fisher Scientific, catalog number: BP164-500)
50× TAE buffer (Thermo Fisher Scientific, catalog number: B49)
SYBRTM Safe DNA gel stain (Thermo Fisher Scientific, catalog number: S33102)
6× gel loading dye (New England BioLabs, catalog number: B7024S)
Nuclease-free water, not DEPC treated (Ambion, catalog number: AM9937)
Ethanol, molecular biology grade
Isopropyl alcohol, molecular biology grade
Custom single-guide RNA (sgRNA 1) TTGGTATTTAGAAAGGTGGT (Synthego)
Custom single-guide RNA (sgRNA 2) GGAAGAAAGAATTGCAATTA (Synthego)
1× TE buffer (included with sgRNA orders from Synthego)
Custom barcoded adapter oligos (IDT)
Note: See Table 1 for a list of barcoded adapter oligos.
Table 1. Barcoded adapters
Barcoded adapter Sequence Barcoded adapter 1 /5Phos/CGCACTCTGATATGTGATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATCACATATCAGAGTGCG Barcoded adapter 2 /5Phos/CTCACAGTCTGTGTGTATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATACACACAGACTGTGAG Barcoded adapter 3 /5Phos/CGCAGCGCTCGACTGTATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATACAGTCGAGCGCTGCG Barcoded adapter 4 /5Phos/TCTGTCTCGCGTGTGTATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATACACACGCGAGACAGA Barcoded adapter 5 /5Phos/CTCTGAGATAGCGCGTATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATACGCGCTATCTCAGAG Barcoded adapter 6 /5Phos/ACACGCGATCTAGTGTATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATACACTAGATCGCGTGT Barcoded adapter 7 /5Phos/ACGCGCGCGTAGTGAGATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATCTCACTACGCGCGCGT Barcoded adapter 8 /5Phos/ACACACGTGTCATGCGATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATCGCATGACACGTGTGT Barcoded adapter 9 /5Phos/ATACTATCTCTCTATGATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATCATAGAGAGATAGTAT Barcoded adapter 10 /5Phos/CACAGTGAGCACGTGAATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGATTCACGTGCTCACTGTG Shrimp alkaline phosphatase (rSAP) (New England BioLabs, catalog number: M0371S or M0371L)
CutSmart® buffer 10× (New England BioLabs, catalog number: B7204S)
Exonuclease III (New England BioLabs, catalog number: M0206S or M0206L)
Cas9 nuclease, S. pyogenes (New England BioLabs, catalog number: M0386T or M0386M)
NEBufferTM 3.1, 10× (New England BioLabs, catalog number: B7203S)
Note: Included with Cas9 Nuclease, S. pyogenes from New England BioLabs.
T4 DNA ligase reaction buffer, 10× (New England BioLabs, catalog number: B0202S)
T4 DNA ligase, HC (Thermo Fisher Scientific, catalog number: EL0013)
SOLu-Trypsin (Sigma-Aldrich, catalog number: EMS0004)
1 kb DNA ladder (carrier DNA) (New England BioLabs, catalog number: N3232S or N3232L)
Recombinant ribonuclease inhibitor (Takara Bio, catalog number: 2313A or 2313B)
Pacific Biosciences® Binding Kits
For Sequel: Sequel Binding and Internal Control kit 3.0 (PacBio, catalog number: 101-626-600)
For Sequel II and IIe: Sequel II Binding kit 2.0 and Internal Control kit 1.0 (PacBio, catalog number: 101-842-900)
Pacific Biosciences® Sequencing Kits
For Sequel: Sequel Sequencing kit 3.0 (PacBio, catalog number: 101-597-900)
Note: Four reactions per kit.
For Sequel II and IIe: Sequel II Sequencing kit 2.0 (PacBio, catalog number: 101-820-200)
Note: Four reactions per kit.
Pacific Biosciences® SMRT® cells
For Sequel: SMRT® Cell 1M v3 tray or SMRT® Cell 1M v3 LR tray (PacBio, catalog number: 101-531-000 or 101-531-001)
For Sequel II and IIe: SMRT Cell 8M tray (PacBio, catalog number: 101-389-001)
AMPure® PB beads (PacBio, catalog number: 100-265-900)
Elution buffer (PacBio, catalog number: 101-633-500)
No-Amp Accessory kit (PacBio, catalog number: 101-788-900)
Sequencing primer v4
10× primer buffer v2
10× annealing buffer
SMRTbell® Enzyme Clean-up kit (PacBio, catalog number: 101-746-400)
Sample plate (PacBio, catalog number: 000-448-888)
Solutions
80% ethanol (see Recipes)
Recipes
80% ethanol
Reagent Final concentration Amount Ethanol (absolute) 80% 40 mL Nuclease-free H2O n/a 10 mL Total n/a 50 mL
Laboratory supplies
15 mL conical tubes (Falcon, catalog number: 352096)
50 mL conical tube (Falcon, catalog number: 1443222)
QIAGEN® genomic tip 100/G (QIAGEN, catalog number: 10243)
QubitTM assay tubes (Thermo Fisher Scientific, catalog number: Q32856)
1.5 mL DNA LoBind tubes (Eppendorf, catalog number: 0030108051)
0.2 mL PCR thermal cycling tubes (Genesee Scientific, catalog number: 27-125)
Equipment
Molecular biology pipettes, standard set
AvantiTM J-15R, IVD, refrigerated benchtop centrifuge (or equivalent) (Beckman Coulter, catalog number: B99517)
Platform shaker
QubitTM quantitation platform (Thermo Fisher Scientific, catalog number: Q33238)
Gel imaging system
Benchtop cooler
Microcentrifuge
Mini centrifuge
Vortex mixer
DynaMagTM-2 magnet (magnetic rack) (Thermo Fisher Scientific, catalog number: 12321D)
PCR thermal cycler
ThermoMixer C with heated lid (or equivalent) (Eppendorf, catalog number: 5382000023)
Sequel, Sequel II, or Sequel IIe Systems (Pacific Biosciences)
Software and datasets
SMRTLink (v12.0, 2023)
ccs (v7.0.0, https://github.com/PacificBiosciences/ccs)
lima (v2.7.1, 2023)
pbmm2 (v1.10.0, 2023)
primrose (v1.2.0, 2022, https://github.com/mattoslmp/primrose)
pb-CpG-tools (v2.3.2, 2023, https://github.com/PacificBiosciences/pb-CpG-tools)
Procedure
文章信息
稿件历史记录
提交日期: May 13, 2024
接收日期: Jul 18, 2024
在线发布日期: Aug 8, 2024
出版日期: Sep 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Tsai, Y. C., Brown, K. A., Bernardi, M. T., Harting, J. and Clelland, C. D. (2024). Single-Molecule Sequencing of the C9orf72 Repeat Expansion in Patient iPSCs. Bio-protocol 14(17): e5060. DOI: 10.21769/BioProtoc.5060.
- Salomonsson, S. E., Maltos, A. M., Gill, K., Aladesuyi Arogundade, O., Brown, K. A., Sachdev, A., Sckaff, M., Lam, K. J. K., Fisher, I. J., Chouhan, R. S., et al. (2024). Validated assays for the quantification of C9orf72 human pathology. Sci Rep. 14(1): 828.
分类
分子生物学 > DNA > DNA 重组
神经科学 > 神经系统疾病
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link