- EN - English
- CN - 中文
Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
拟南芥子房中轴与侧区的激光辅助显微切割及高通量RNA测序
发布: 2024年09月05日第14卷第17期 DOI: 10.21769/BioProtoc.5056 浏览次数: 2063
评审: Samik BhattacharyaShengze Yao

相关实验方案
对拟南芥样本的cDNA 3'末端进行快速扩增(3' RACE)
Encarnación Rodríguez-Cazorla [...] Antonio Vera
2015年10月05日 21342 阅读
Abstract
For obtaining insights into gene networks during plant reproductive development, having transcriptomes of specific cells from developmental stages as starting points is very useful. During development, there is a balance between cell proliferation and differentiation, and many cell and tissue types are formed. While there is a wealth of transcriptome data available, it is mostly at the organ level and not at specific cell or tissue type level. Therefore, methods to isolate specific cell and tissue types are needed. One method is fluorescent activated cell sorting (FACS), but it has limitations such as requiring marker lines and protoplasting. Recently, single-cell/nuclei isolation methods have been developed; however, a minimum amount of genetic information (marker genes) is needed to annotate/predict the resulting cell clusters in these experiments. Another technique that has been known for some time is laser-assisted microdissection (LAM), where specific cells are microdissected and collected using a laser mounted on a microscope platform. This technique has advantages over the others because no fluorescent marker lines must be made, no marker genes must be known, and no protoplasting must be done. The LAM technique consists in tissue fixation, tissue embedding and sectioning using a microtome, microdissection and collection of the cells of interest on the microscope, and finally RNA extraction, library preparation, and RNA sequencing. In this protocol, we implement the use of normal slides instead of the membrane slides commonly used for LAM. We applied this protocol to obtain the transcriptomes of specific tissues during the development of the gynoecium of Arabidopsis.
Key features
• Laser-assisted microdissection (LAM) allows the isolation of specific cells or tissues.
• Normal slides can be used for LAM.
• It allows the identification of the transcriptional profiles of specific tissues of the Arabidopsis gynoecium.
Keywords: Laser-assisted microdissection (激光辅助显微切割)Graphical overview
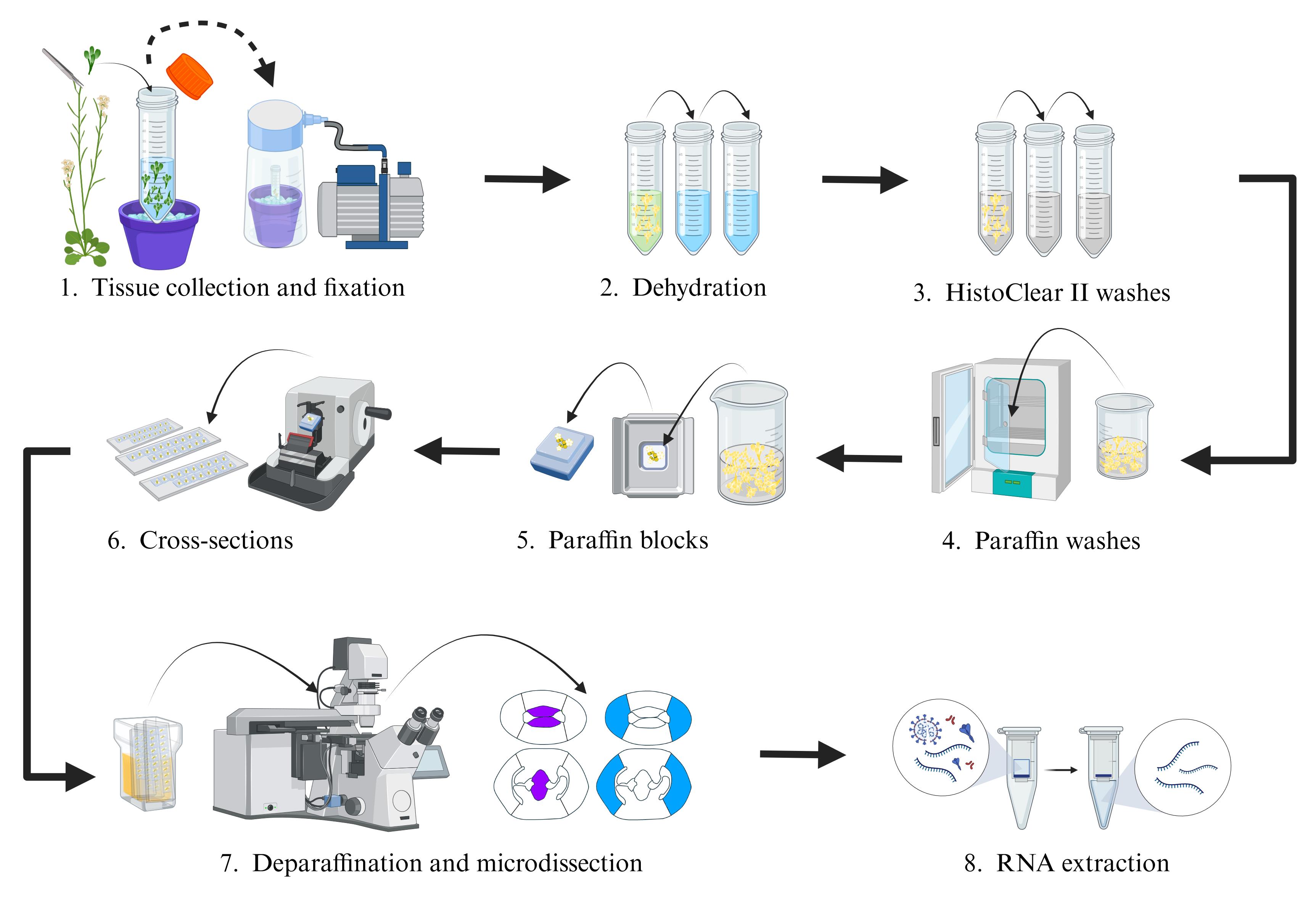
Laser-assisted microdissection (LAM) procedure of gynoecium tissues in Arabidopsis
Background
Multicellular organisms, such as animals and plants, are made up of many cells, which are grouped together to form different tissues and organs. Proper development requires complex molecular regulation, ranging from changes in gene expression and signaling events to hormonal activity. Despite the efforts made to understand these mechanisms that regulate development, the great morphological complexity of tissues or organs represents a limitation to creating tissue-specific expression profiles [1–4]. However, in recent years, several techniques have been successfully developed and implemented to isolate and characterize the transcriptomic profiles of specific cell populations, such as fluorescent activated cell sorting (FACS), single-cell isolation, single-nuclei isolation, and laser-assisted microdissection (LAM) [5]. LAM is a microscopy-based technique that has established itself as a good technique for isolating and collecting specific cell populations from their tissue context [6–11]. The LAM technique is distinguished from others for not requiring the generation of marker lines or protoplasts or previous information on a large number of marker genes; it only requires knowing the morphology of the tissues to be collected. These characteristics have allowed expanding LAM implementation to different organs of different plant species such as tomato fruit [12, 13], rice [14], California poppy [15], and Arabidopsis [16–18]. Here, we describe a protocol, built on previous work by Chávez Montes et al., 2016 [10], to perform LAM and RNA-seq of four tissue types from the Arabidopsis gynoecium: carpel marginal meristem (CMM) and septum (SEP), both tissues of the medial domain of the gynoecium, and the presumptive carpel wall (PC) and valves (VV), two tissues that make up the lateral domain of the gynoecium [19]. The aim was to identify the transcriptomic profiles that regulate their development. In the implementation of this protocol, the use of slides without the membrane was chosen instead of the commercially available slides (MembraneSlides) for LAM, which was better for the collection of the tissues of interest. Furthermore, we briefly describe the data analysis and gene expression visualization. This protocol should also be applicable for other plant species.
Materials and reagents
Biological materials
Arabidopsis thaliana plants (inflorescence tissue)
Reagents
Absolute ethyl alcohol for molecular biology (ethanol) (Karal, catalog number: 2003)
Glacial acetic acid (Fermont, catalog number: 03015)
Histo-Clear II (Electron Microscopy Sciences, catalog number: 64111-04)
Paraplast Plus® (paraffin) (Sigma-Aldrich, catalog number: P3683)
Methyl alcohol (Karal, catalog number: 2010)
Direct-zol RNA Microprep (Zymo Research, catalog number: R2063)
Solutions
Fixation solution (see Recipes)
Note: We do not recommend using formaldehyde (FDA; paraformaldehyde) as the fixation solution, because a cross-link reversal step would be needed for RNA extraction. Some discussion on fixation solutions is given in Chávez Montes et al. [10].
Recipes
Fixation solution
Reagent Final concentration Quantity or Volume Ethanol (absolute) 75% (v/v) 3 mL Glacial acetic acid 25% (v/v) 1 mL Total n/a 4 mL
Laboratory supplies
Sterile falcon tubes
Beakers
Dissecting forceps
Steel inclusion molds 15 mm × 15 mm (Citotest)
Embedding cassettes without lid, color white (Simport Scientific, catalog number: M480-2)
Microscope slides 26 × 76 mm, thickness of 1 mm (LAUKA)
Low-profile disposable blades (Leica, model: 819)
Slide rack and glass staining dish with lid (LaboLan, catalog number: 29600001XL)
TubeCollector 2×200 CM II (Zeiss, catalog number: 415101-2000-410)
AdhesiveCap 200 clear (Zeiss, catalog number: 415190-9191-000)
MembraneSlide 1.0 OPEN (Zeiss, catalog number: 415101-4401-000)
Equipment
Vacuum pump (Gast, model: DOA-P704-AA)
Vacuum desiccator
Mini-rotator, 2–80 rpm w/disk & clamps (Glas-Col, catalog number: 099A MR1512)
Convection incubator (Binder, model: BD 23)
Paraffin dispenser (Leica, model: EG 1120)
Microtome (Leica, model: RM2035)
Slide warmer (Lab-Line Instruments, model: 26005)
PALM MicroBeam IV microscope with motorized RoboMover and CapMover (Zeiss, series number: 1214000156)
NanoDrop (optional)
Covaris S2 ultrasonicator (optional)
Qubit 4 fluorometer (optional)
Bioanalyzer 2100 (optional)
Software and datasets
PALM Robo software (Zeiss, v4)
Optional: R studio for RNA-seq data analysis
Procedure
文章信息
稿件历史记录
提交日期: May 9, 2024
接收日期: Jul 22, 2024
在线发布日期: Aug 2, 2024
出版日期: Sep 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Luna-García, V. and de Folter, S. (2024). Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains. Bio-protocol 14(17): e5056. DOI: 10.21769/BioProtoc.5056.
- Luna-García, V., Bernal Gallardo, J. J., Rethoret-Pasty, M., Pasha, A., Provart, N. J. and de Folter, S. (2024). A high-resolution gene expression map of the medial and lateral domains of the gynoecium of Arabidopsis. Plant Physiol. 195(1): 410–429.
分类
植物科学 > 植物分子生物学 > RNA > 转录
分子生物学 > RNA > 转录
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link











