- EN - English
- CN - 中文
Protocol for Imaging the Same Class IV Neurons at Different Stages of Development
在不同发育阶段成像同一类IV型神经元的操作步骤
发布: 2024年08月20日第14卷第16期 DOI: 10.21769/BioProtoc.5052 浏览次数: 1317
评审: Nafisa M. JadavjiEhsan KheradpezhouhAnonymous reviewer(s)
Abstract
In this protocol, we focused on analyzing internal branches of Drosophila class IV neurons. These neurons are characterized by their highly branched axons and dendrites and intricately tile the larval body. As Drosophila larvae progress through developmental stages, the dendritic arbors of Class IV neurons undergo notable transformations. As Drosophila larvae develop, their Class IV dendritic arbors grow. In the initial 24 h after egg laying (AEL), the dendrites are smaller than segments. During the subsequent 24 h of the first instar larval stage, dendritic arbors outpace segment growth, achieving tiling. After 48 h, arbors and segments grow concurrently. Epidermal cells near Class IV dendrites expand in proportion to segment growth. This observation suggested that Class IV cells might grow via branch dilation—uniformly elongating branches, akin to Class I cells [1,2]. To understand whether the class IV complex arbor structure is formed by dilation or simply from growing tips, we developed this protocol to introduce a systematic approach for quantitatively assessing the growth dynamics of internal branches.
Key features
• This protocol employs imaging the same neuron over different development times
• Drosophila embryo and larvae genotype is ;;ppkCD4-tdGFP, which explicitly tags class IV neurons
• This protocol for the preparation of agar pads to mount and image Drosophila larvae is adapted from Monica Driscoll's method
• Neurons are imaged without the use of anesthetics and for a short duration of time
• This technique involves the use of a spinning disk confocal microscope
Keywords: Class IV neurons (IV型神经元)Graphical overview
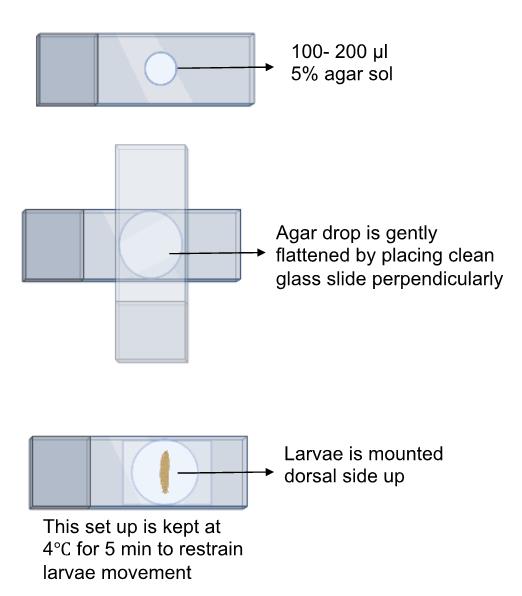
Schematic overview of the process for mounting larvae on an agar pad for imaging using a confocal microscope
Background
Dendrites serve as extensions of neurons and play a pivotal role in receiving signals from other neurons or the surrounding environment. These extensions often have a branched structure, which enhances their capacity to gather information. An excellent model for exploring the development of dendrites is found in the extensively branched Drosophila Class IV neurons. These neurons exhibit a wide array of shapes in their dendritic structures, with each dendrite being unique. This suggests that the growth of dendrites is more likely a random process rather than a predetermined one.
Despite this diversity, the specific underlying mechanisms guiding the development of dendrites in these neurons remain largely unclear. The larval body is divided into segments, and each segment harbors a pair of class IV neurons. These neurons, in our hand, are first visible around 16 h after egg laying (AEL). At this stage, neurons are small as compared to their harboring segment, but they grow fast and catch up with growing segments. After that, neurons grow in proportion to the expansion of the segments [3]. This has led to the hypothesis that Class IV cells expand through branch dilation—that is, by uniform expansion of branches along their lengths [4]. Such dilation occurs in Class I cells [1]. To form the highly branched late-stage dendrite, dilation would need to be complemented by the formation of new branches on extant ones, to infill the structure.
Through our investigation, we have made a noteworthy observation: dendritic branches lengthen primarily through extension from their tips, rather than by expanding their non-terminal branches. We developed this protocol to image the same neurons over developmental time without the use of anesthetics. Our focus was to image neurons for short durations and take static images, placing larvae back in apple agar plates in the Darwin chamber at 25 °C so that neurons and larvae do not get stressed by continuous imaging and use of anesthetics. The LarvaSPA method [5] allows imaging neurons continuously for 10 h; however, our goal was to image the same neuron every 24 h at various developmental time points.
Materials and reagents
Biological material
Fly line ;;ppk-cd4-tdGFP (homozygous) was used to image class IV dendritic arborization neurons and was a gift from C. Han (Cornell University)
Apple juice unsweetened frozen concentrate (4×) [Stop & Shop (grocery store), 12 oz can]
Dextrose (Sigma-Aldrich, catalog number: G5767)
Sucrose (Sigma-Aldrich, catalog number: 57-50-1)
Bacto agar (Becton Dickinson, catalog number: 214010)
NaOH pellet (Sigma-Aldrich, catalog number: 221465)
Propionic acid (Sigma-Aldrich, catalog number: 402907)
Phosphoric acid (Sigma-Aldrich, catalog number: 695017)
Halocarbon oil 700 (Sigma-Aldrich, catalog number: H8898)
Agarose (AmericanBio, catalog number: AB9012-36-6)
PBS 10× solution (AmericanBio, catalog number: AB11072-01000)
High-precision microscope cover glass 22 × 22 mm (VWR International, catalog number: 16004-302)
35 mm dish, No. 1.5 coverslip 10 mm glass diameter uncoated (MatTex, catalog number: P35G-1.5-10-C)
Mini embryo collection cages (Flystuff.com) (Genesee Scientific, catalog number: 59-105)
Tissue culture dish, 35 × 10 mm style (Falcon, catalog number: 353001)
Tray of 100 NARROW 1-oz PS vials, each with 10 mL of glucose media, pre-loaded with cellulose-acetate plugs (Archon Scientific, catalog number: D20102)
Frosted micro slides (VWR, catalog number: 48312-004)
Equipment
For imaging, samples were mounted on the microscope stage, illuminated with Nikon lasers (488 nm at 18%–21% laser power), and imaged on a spinning disk microscope: Yokogawa CSU-W1 disk (pinhole size 50 μm) built on a fully automated Nikon TI inverted microscope with perfect focus system, an sCMOS camera (Zyla 4.2 plus sCMOS), and Nikon Elements software with either a 40× (1.25 NA) or 20× (0.5 NA) water immersion objective.
Software and datasets
Images were stitched and analyzed using Fiji (NIH, 2021). Detailed procedure of stitching is mentioned in the procedure section. https://imagej.net/software/fiji/
NIS-Elements Imaging Software (Nikon, 2022 This imaging software comes with the spinning disk microscope. It is an interface that enables the user to capture images using Nikon microscope and also helps to export the raw images in the TIFF format, which could later be processed by FIJI for stitching
Prism 9 (GraphPad Prism, 2023). https://www.graphpad.com/scientific-software/prism/
Procedure
文章信息
稿件历史记录
提交日期: Jun 30, 2024
接收日期: Jul 12, 2024
在线发布日期: Aug 2, 2024
出版日期: Aug 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Shree, S. and Howard, J. (2024). Protocol for Imaging the Same Class IV Neurons at Different Stages of Development. Bio-protocol 14(16): e5052. DOI: 10.21769/BioProtoc.5052.
- Shree, S., Sutradhar, S., Trottier, O., Tu, Y., Liang, X. and Howard, J. (2022). Dynamic instability of dendrite tips generates the highly branched morphologies of sensory neurons. Sci Adv. 8(26): eabn0080. https://doi.org/10.1126/sciadv.abn0080
分类
发育生物学 > 形态建成 > 细胞结构
神经科学 > 细胞机理 > 神经元命运
细胞生物学 > 细胞成像 > 共聚焦显微镜
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link