- EN - English
- CN - 中文
Extraction of Bacterial Membrane Vesicle and Phage Complex by Density Gradient Ultracentrifugation
通过密度梯度超速离心法提取细菌膜泡和噬菌体复合物
(*contributed equally to this work) 发布: 2024年08月20日第14卷第16期 DOI: 10.21769/BioProtoc.5050 浏览次数: 2032
评审: Alba BlesaNuttavut KosemManousos E. Kambouris

相关实验方案
诱导型HIV-1库削减检测(HIVRRA):用于评估外周血单个核细胞中HIV-1潜伏库清除策略毒性与效力的快速敏感方法
Jade Jansen [...] Neeltje A. Kootstra
2025年07月20日 2457 阅读
Abstract
The bacterial membrane vesicles (MVs) are non-replicative, nanoscale structures that carry specific cargos and play multiple roles in microbe–host interactions. An appropriate MV isolation method that mimics complex pathogen infections in vivo is needed. After bacterial MVs extraction, flagella or pili can be frequently observed along with MVs by transmission electron microscope (TEM). Recently, MVs from Pseudomonas aeruginosa were found to coexist with Pf4 phages, and this MV–phages complex exhibited a different impact on host cell innate immunity compared with MVs or phages solely. The presence of this MVs–phages complex simulates the real condition of complex pathogen infections within the host. This protocol outlines the extraction of the MVs and Pf4 phages complex of P. aeruginosa PAO1, including the respective isolation and qualification approaches. Our step-by-step bacterial MVs–phages complex extraction protocol provides valuable insights for further studying microbe–host cell interactions and the development of novel phage therapies.
Key features
• Detailed density gradient extraction procedures of MVs–phages complex
• TEM, plaque assay, and PCR to verify the coexistence of MVs and phages
• The obtained MVs–phages complex can be used for exploring phage–microbe–host cell interactions
Keywords: Pseudomonas aeruginosa (铜绿假单胞菌)Graphical overview
Background
Pseudomonas aeruginosa is an opportunistic pathogen that often causes serious hospital-acquired infections and can lead to a variety of infectious diseases including pneumonia, wound infections, and septicemia [1]. Due to the high mortality rate of these infections, P. aeruginosa was recognized as one of the most life-threatening bacteria and listed as a priority pathogen for research and development of new antibiotics by the World Health Organization [2].
The Pf phage, a filamentous inovirus specifically associated with P. aeruginosa, is a type of temperate phage, which can be classified from Pf1 to Pf8. Approximately half of P. aeruginosa isolates harbor Pf phage operons within their genomes [3,4]. Pf phages are about 6–7 nm in diameter and vary in length from 0.8 to 2 µm, with a single-stranded DNA (ssDNA) as the genetic material packaged within a helical filamentous structure major coat protein CoaB [3]. Of these, Pf4 phage is an important member, and its genome is integrated into the P. aeruginosa PAO1 as the prophage. Thus, Pf4 phage can emerge inside the bacteria [5]. The core genes of the Pf4 phages mainly include xisF4, PA0720, PA0723, PA0724, and PA0726, which encode excisionase, single-stranded DNA binding protein, major coat protein CoaB, minor coat protein CoaA, and morphogenesis protein, respectively [3,5]. A recent study showed that Pf4 phages can be transmitted between bacteria and have various effects on P. aeruginosa, including reducing twitching motility, altering biofilm production, and affecting the release of virulence factors, which are important in the pathogenesis of P. aeruginosa [6–8]. However, how Pf4 is transmitted has remained elusive.
Bacterial membrane vesicles (MVs) are structures produced by bacteria with a diameter ranging from 40 to 400 nm that carry specific cargo and travel a long distance (centimeter scale) to play essential roles in cell–cell communications [9,10]. Gram-negative bacteria MVs could be divided into two types: B (blebbing)-type vesicles, which are formed via blebbing from the outer membrane, and E (explosive)-type vesicles, which are formed due to explosive cell lysis [11,12]. Studies have suggested that P. aeruginosa E-MVs can be formed under certain stresses and bacteriophage lysis conditions induced by Pf4 phages [3].
Many previous works observed that after the crude MV extraction step, flagella can also be observed by TEM [13]. A recent study investigated the combined effects of P. aeruginosa MVs with phages upon the recognition function of the host’s innate immune system. In a mouse model of acute lung infection, treatment with MVs-Pf4 phages complex resulted in reduced infiltration of neutrophils in the lung alveoli, thus suppressing the occurrence of inflammatory reactions [3].
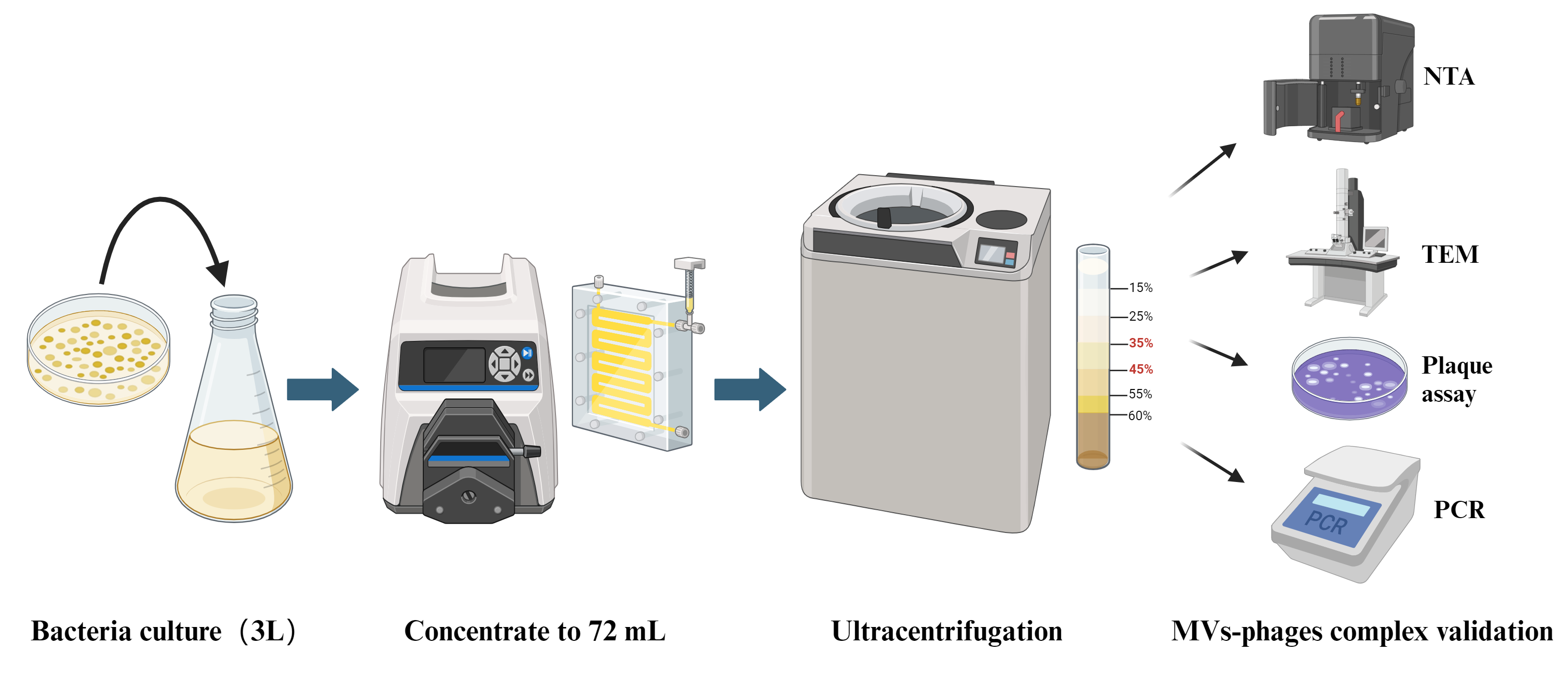
In this protocol, we establish a bacterial MVs–phages co-extraction platform from P. aeruginosa PAO1 as a model organism. The MVs–phages extract effectively mimics the complex MVs–phage coexistence condition in vivo within the host during pathogen infections. Overall, this is an optimized method for extracting the MV–phage complex based on density gradient centrifugation (Figure 1), with feasibility verified through TEM, PCR, and plaque assays. It will provide a reliable protocol for studying bacteriophage–microbe–host cell interactions.
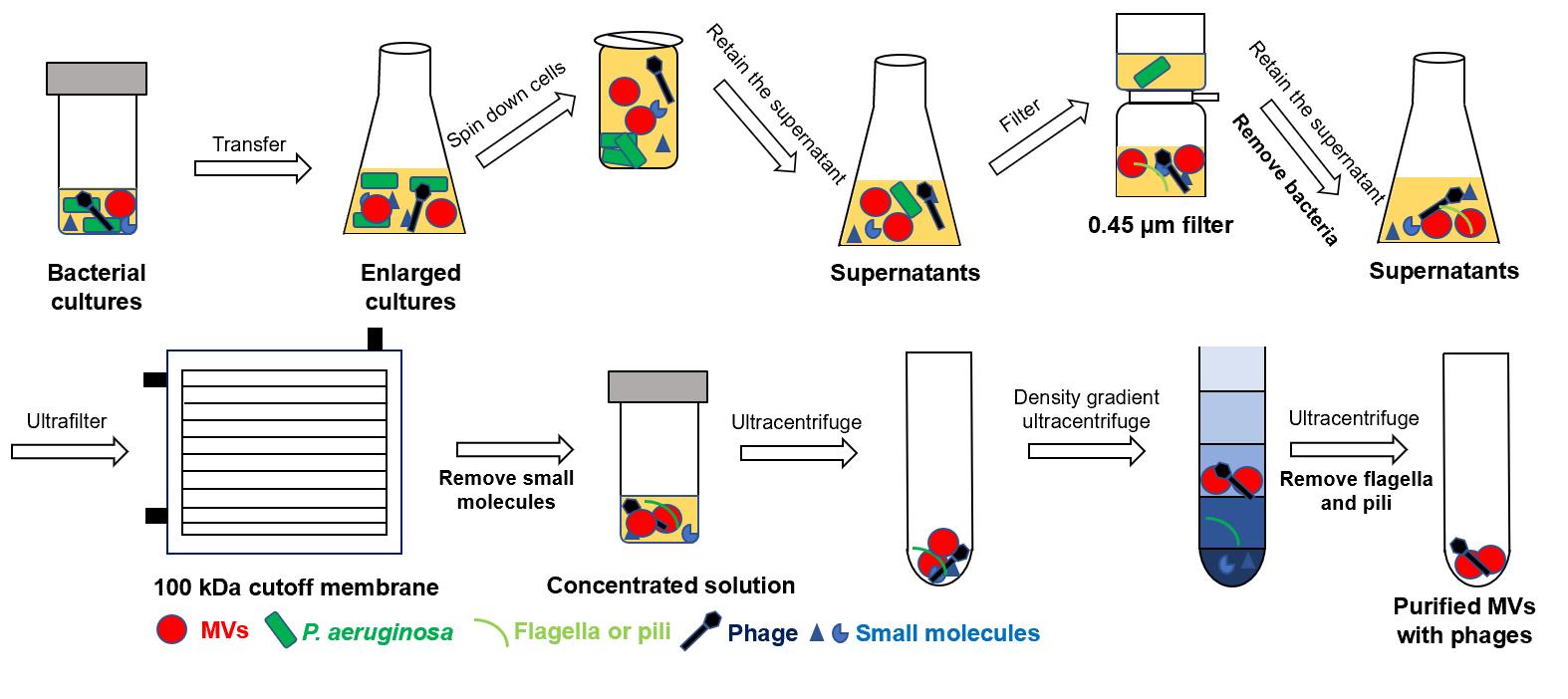
Figure 1. Flowchart of MV–phage complex extraction. The bacterial culture is centrifuged to retain the supernatant. After filtering and ultrafiltering, bacteria and small molecules can be removed. After density gradient ultracentrifuging, flagella and pili can be removed. Then, purified MVs–phage samples can be obtained.
Materials and reagents
The following materials and equipment are recommended for this protocol, but alternatives from other sources can also be used when proven to be equivalent.
Biological materials
P. aeruginosa PAO1 strain (strains are preserved at -80 °C in our laboratory)
P. aeruginosa PAO1 ΔPf4 strain (strains are preserved at -80 °C in our laboratory)
Opti-Prep gradient medium (Sigma-Aldrich, catalog number: D1556)
Phosphate-buffered saline (PBS) (Servicebio, catalog number: G4202)
2% Phosphotungstic acid solution (Acmec, catalog number: AG1599)
LB liquid medium (Servicebio, catalog number: G3103)
Premix TaqTM Version 2.0 (Takara, catalog number: R004Q)
1% agarose gel (Baygene, catalog number: 192255)
DNA marker (Coolaber, catalog number: D2000)
Pancreatic digest of casein (peptone) (Beyotime, catalog number: ST800)
Soybean digest (Sigma-Aldrich, catalog number: C7210)
NaCl (Dogesce, catalog number: Sodium Chloride)
Agar (Dogesce, catalog number: Agar)
CaCl2 (Macklin, catalog number: C915443-500g)
MgCl2 (Macklin, catalog number: M813766-500g)
Mitomycin C (AbMole, catalog number: M5791)
GelRed 10,000× gel staining solution (I-PRESCI, catalog number: P202-01)
TSB culture medium (see Recipes)
1.5% LB agar solid medium (see Recipes)
0.75% LB agar semi-solid medium (see Recipes)
TSB culture medium
The composition of the medium includes distilled water, 1.5% pancreatic digest of casein (peptone), 0.5% soybean digest, and 0.5% NaCl.
1.5% LB agar solid medium
100 mL LB liquid medium with 1.5 g of agar.
0.75% LB agar semi-solid medium
100 mL LB liquid medium with 0.75 g agar, 1 mM CaCl2, and 1 mM MgCl2.
Laboratory supplies
LB agar plate (Servicebio, catalog number: G3104-0910)
50 mL cell culture tube (Bioland, catalog number: ATS05-12-50)
13.2 mL ultracentrifuge tube (Beckman, catalog number: 344059)
1.5 mL centrifuge tube (Eppendorf, catalog number: 0030108051)
PCR tube (Eppendorf, catalog number: 0030124359)
500 mL filter flask with 0.45 μm filter (Biosharp, catalog number: BS-500-XT)
Glow-discharged 200-mesh carbon grid (Beijing Zhongjingkeyi Technology, catalog number: BZ11022B)
250 mL centrifuge bottle (Beckman, catalog number: 356011)
Equipment
Intelligent high-efficiency centrifuge (Avanti, model: JXN-26)
Masterflex easy-load machine (Sartorius AG, model: VFA013)
100 KD Vivaflow membranes (Sartorius AG, model: VF20H4)
Ultracentrifuge machine (Beckman, model: Optima XE-100)
High-pressure sterilization pot (Yamato, model: SQ810C)
Constant temperature shaker (Shanghaiminquanyiqi, model: MQT-60R)
Tecan Spark 10 M plate reader (Tecan Group Ltd., model: Tecan Spark 10 M)
Transmission electron microscope (Hitachi, model: HT7700)
Nano Sight nanoparticle tracking analysis machine (Malvern Instruments Ltd, model: NS300)
Procedure
文章信息
稿件历史记录
提交日期: May 9, 2024
接收日期: Jul 12, 2024
在线发布日期: Aug 2, 2024
出版日期: Aug 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Li, S., Ren, A., Li, M., Li, G., Yang, L. and Jia, T. (2024). Extraction of Bacterial Membrane Vesicle and Phage Complex by Density Gradient Ultracentrifugation. Bio-protocol 14(16): e5050. DOI: 10.21769/BioProtoc.5050.
分类
微生物学 > 微生物细胞生物学 > 细胞器分离
微生物学 > 微生物-宿主相互作用 > 病毒
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link