- EN - English
- CN - 中文
Determination of Ligand-Target Interaction in vitro by Cellular Thermal Shift Assay and Isothermal Dose-response Fingerprint Assay
通过细胞热转移法和等温剂量反应指纹分析法测定体外配体-靶标相互作用
发布: 2024年08月05日第14卷第15期 DOI: 10.21769/BioProtoc.5047 浏览次数: 4911
评审: Dipak Kumar PoriaSrajan KapoorGundeep KaurAnonymous reviewer(s)

相关实验方案
利用近端连接分析在新鲜分离的大鼠血管平滑肌细胞中定量Kv7.4与Dynein蛋白的相互作用
Jennifer van der Horst and Thomas A. Jepps
2024年03月20日 1563 阅读
Abstract
The cellular thermal shift assay (CETSA) and isothermal dose-response fingerprint assay (ITDRF CETSA) have been introduced as powerful tools for investigating target engagement by measuring ligand-triggered thermodynamic stabilization of cellular target proteins. Yet, these techniques have rarely been used to evaluate the thermal stability of RNA-binding proteins (RBPs) when exposed to ligands. Here, we present an adjusted approach using CETSA and ITDRFCETSA to determine the interaction between enasidenib and RBM45. Our assay is sensitive and time-efficient and can potentially be adapted for studying the interactions of RBM45 protein with other potential candidates.
Key features
• This protocol builds upon the method developed by Molina et al. and extends its application to new protein classes, such as RBPs.
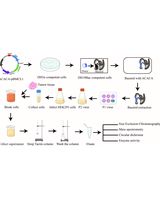
Keywords: Ligand–target interaction (配体-靶标相互作用)Graphical overview
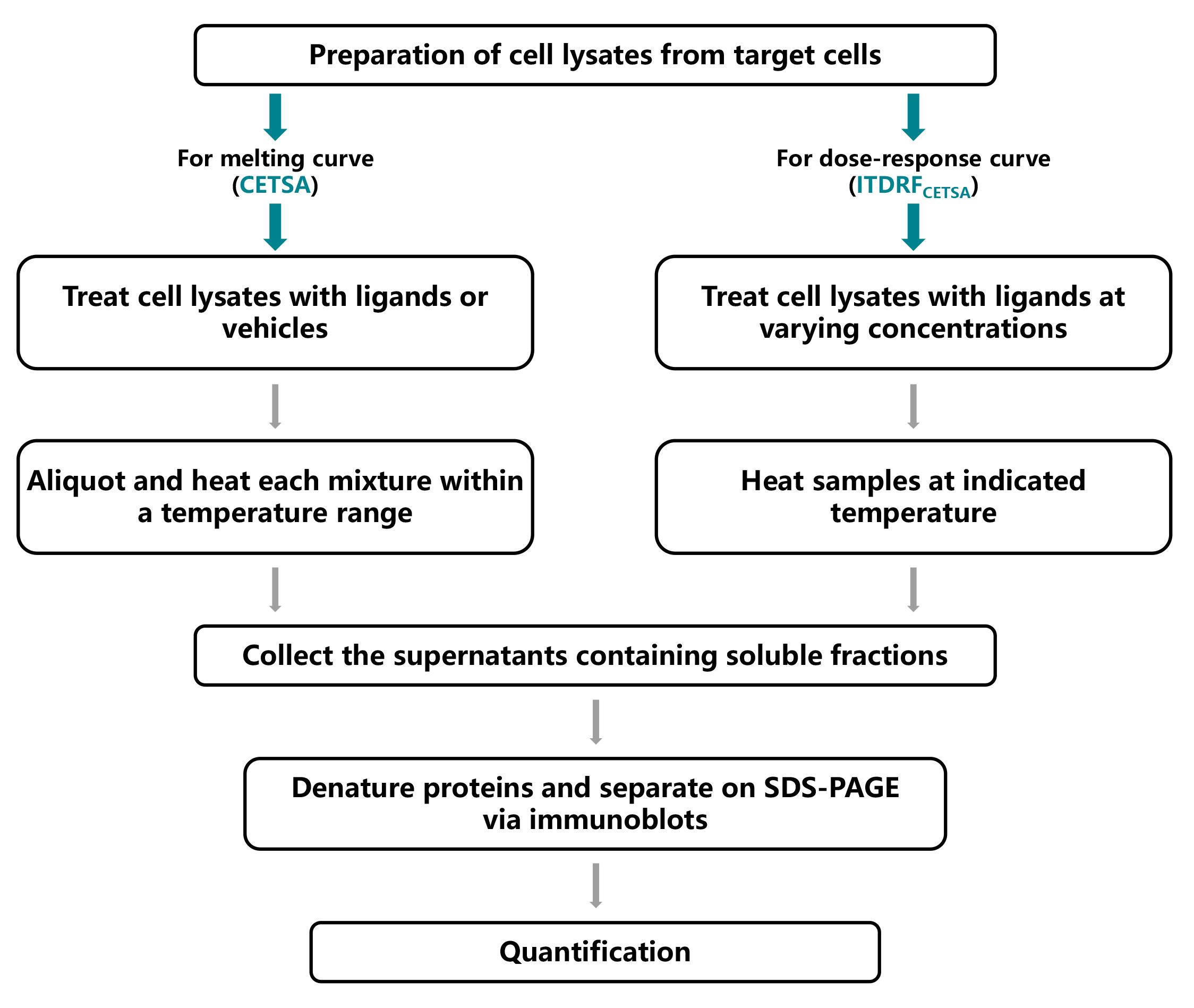
Background
RNA-binding proteins (RBPs) contribute considerably to post-transcriptional regulation; thus, their dysregulation has been linked to a variety of illnesses, including malignancies [1]. Thereafter, targeting RBPs with specific ligands to perturb cancer development has attracted much attention [2]. Recent studies, including ours, have unraveled the role of RNA-binding motif protein 45 (RBM45) in promoting hepatocellular carcinoma (HCC) progression [3,4], highlighting its importance in cancer development. More crucially, our previous work, which combines a molecular docking approach as well as molecular and cellular techniques, identified enasidenib, a previously considered mutant isocitrate dehydrogenase 2 (mIDH2) inhibitor that binds to RBM45. Therefore, a better clarification of the binding affinity between enasidenib and RBM45 would be beneficial for the design of RBM45-targeted therapeutics.
Recently, several label-free techniques have been developed to determine ligand-target engagement using intact cells or cell lysates. These techniques include drug affinity responsive target stability (DARTS) [5], stability of proteins from rates of oxidation (SPROX) [6], and cellular thermal shift assay (CETSA) [7]. Among these, CETSA is more widely applicable since it can determine the specific ligand-induced target thermodynamic stabilization in biological settings such as whole cell lysates, intact cells, or even tissues [7]. Typically, the shift in thermal stability is assessed by quantifying the density of the remaining soluble target proteins using available antibodies via immunoblots analysis. As a result, thermal melt curves are generated, in which the target protein of interest, with or without ligands, is exposed to a panel of temperatures in the CETSA experiment. Alternatively, an isothermal dose-response curve is also generated, in which the target protein is subjected to increasing concentrations of compounds under a constant temperature (a checkpoint at which the unliganded target protein is degraded as determined by the CETSA experiment), referred to as the isothermal dose-response fingerprint assay (ITDRFCETSA) [8]. Undoubtedly, CETSA and ITDRFCETSA techniques offer the opportunity to investigate whether ligands engage their intended targets in complicated environments and determine at what concentration they exert their effects.
Moreover, the CETSA approach has been demonstrated to be effective in determining the thermal stability of enzymes such as dihydrofolate reductase (DHFR) and thymidylate synthase (TS) [7], kinases including cyclin-dependent kinases CDK4 and CDK6 [7], mitogen‐activated protein kinases p38α and ERK1/2 [8]), as well as membrane proteins like solute carriers (SLCs) [9], in the absence and presence of potential ligands. However, the CETSA technique has rarely been applied to assess the thermodynamic stabilization of RBPs when subjected to compounds. Furthermore, a cell-based CETSA decreases the sensitivity to the effects of low-affinity ligands due to drug dissociation from the target after cell lysis, making lysate-based CETSA a preferred choice [9]. Here, we report the modified cell lysate CETSA and ITDRFCETSA techniques for determining the interaction between enasidenib and RBM45. Our assay is sensitive and time saving and can potentially be adapted to investigate RBM45 protein with other candidate ligands.
Materials and reagents
Cell line
Human liver cancer cell line SK-HEP-1 was purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China).
Materials
1.5 mL tubes (Corning Axygen, catalog number: MCT-150-C)
200 μL tubes (Corning Axygen, catalog number: PCR-02-C)
10 cm dishes (Corning Axygen, catalog number: 430167)
Reagents
MEM medium (Thermo Fisher Scientific, catalog number: 61100061)
Fetal bovine serum (FBS) (Royacel SERUM, catalog number: RYS-KF22-01)
Non-essential amino acids (100×) (Thermo Fisher Scientific, catalog number: 11140050)
Sodium pyruvate 100 mM solution (Thermo Fisher Scientific, catalog number: 11360070)
Penicillin-Streptomycin (100×) (New Cell & Molecular Biotech Co., Ltd., catalog number: C100C5)
1× phosphate buffered saline (PBS), pH 7.4 (Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd, catalog number: ZQ-1300)
0.25% trypsin-EDTA (New Cell & Molecular Biotech Co., Ltd., catalog number: C125C1)
RIPA lysis buffer (Beyotime Biotechnology, catalog number: P0013B)
ProtLytic protease inhibitor cocktail (EDTA-Free, 100× in DMSO) (New Cell & Molecular Biotech Co., Ltd., catalog number: P001)
Liquid nitrogen
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: 67-68-5)
Enasidenib (Jiangsu Aikon Biopharmaceutical R&D Co., Ltd., catalog number: 4409639)
Bicinchoninic acid (BCA) Protein Assay kit (enhanced) (Beyotime Biotechnology, catalog number: P0009)
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, 5× (Beyotime Biotechnology, catalog number: P0015)
Rabbit-polyclonal-anti-RBM45 (ABclonal, catalog number: A13843)
Goat anti-Rabbit IgG (H+L) secondary antibody, HRP (Thermo Fisher Scientific, catalog number: 31460)
Enhanced chemiluminescence (ECL) detection reagent (Vazyme, catalog number: E411-03/04/05)
Equipment
Gene amplification instrument (Bioer Technology, model: G1000 GeneExplorer)
Metal Bath (Labnet)
Electrophoresis systems & transfer equipment (Bio-Rad)
Image analyzer (Tanon, model: 5200)
Refrigerated high-speed benchtop centrifuge (Thermo Fisher Scientific, model: 75005289)
Rotating incubator (Haimen Kylin-Bell Lab Instruments Co., Ltd., model: QB-228)
Software and datasets
ImageJ 1.46r
Microsoft Excel
GraphPad Prism 9.0.0
Procedure
文章信息
稿件历史记录
提交日期: Jan 12, 2024
接收日期: Jun 28, 2024
在线发布日期: Jul 19, 2024
出版日期: Aug 5, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Du, D., Yuan, S. and Xiong, J. (2024). Determination of Ligand-Target Interaction in vitro by Cellular Thermal Shift Assay and Isothermal Dose-response Fingerprint Assay. Bio-protocol 14(15): e5047. DOI: 10.21769/BioProtoc.5047.
分类
分子生物学 > 蛋白质 > 蛋白质-蛋白质相互作用
癌症生物学 > 癌症生物化学 > 蛋白质
药物发现 > 药物设计
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link