- EN - English
- CN - 中文
An NMR Approach for Investigating Membrane Protein–Lipid Interactions Using Native Reverse Micelles
利用天然反向胶束的核磁共振方法研究膜蛋白-脂质相互作用
发布: 2024年07月20日第14卷第14期 DOI: 10.21769/BioProtoc.5039 浏览次数: 2267
评审: Qingliang ShenMarc-Antoine SaniAnonymous reviewer(s)
Abstract
Peripheral membrane proteins (PMPs) are a subgroup of membrane-associated proteins that are water-soluble and bind to membranes, often reversibly, to perform their function. These proteins have been extensively studied in the aqueous state, but there is often a lack of high-resolution structural and functional studies of these proteins in the membrane-bound state. Currently, nuclear magnetic resonance (NMR) is among the best-equipped methods to study these relatively small proteins and domains, but current models have some disadvantages that prevent a full understanding of PMP interactions with membranes and lipids. Micelles, bicelles, and nanodiscs are all available for NMR observation but are based on synthetic lipids that may destabilize proteins or are too large to accommodate straightforward structural analysis. This protocol introduces a method for forming reverse micelles using lipids from natural sources, here called native reverse micelles. This technique allows the PMPs to embed within a shell of naturally derived lipids surrounding a small water core solubilized in an alkane solvent. PMP embedment in the lipid shell mimics binding to a cellular membrane. Here, naturally derived lipids from soy, bovine heart, and porcine brain are used in conjunction with n-dodecylphosphocholine (DPC) to encapsulate a PMP from either concentrated or dried protein, resulting in reverse micelles that may be confirmed via dynamic light scattering and NMR. This protocol allows for high-quality NMR data of PMPs interacting with membrane lipids within a biologically accurate environment.
Key features
• This protocol describes using natural lipids to construct reverse micelles for high-resolution NMR studies of proteins.
• Initial optimization of encapsulation conditions proceeds through visual assessment, with dynamic light scattering (DLS) to measure size distribution, and NMR to observe protein behavior.
• Membrane-interacting proteins are encapsulated in their membrane-bound state. Proteins that do not interact with membranes are housed in their water-solubilized state.
• Structural, functional, and inhibitory studies may be performed on native reverse micelle-encapsulated proteins.
Keywords: Membrane models (膜模型)Graphical overview
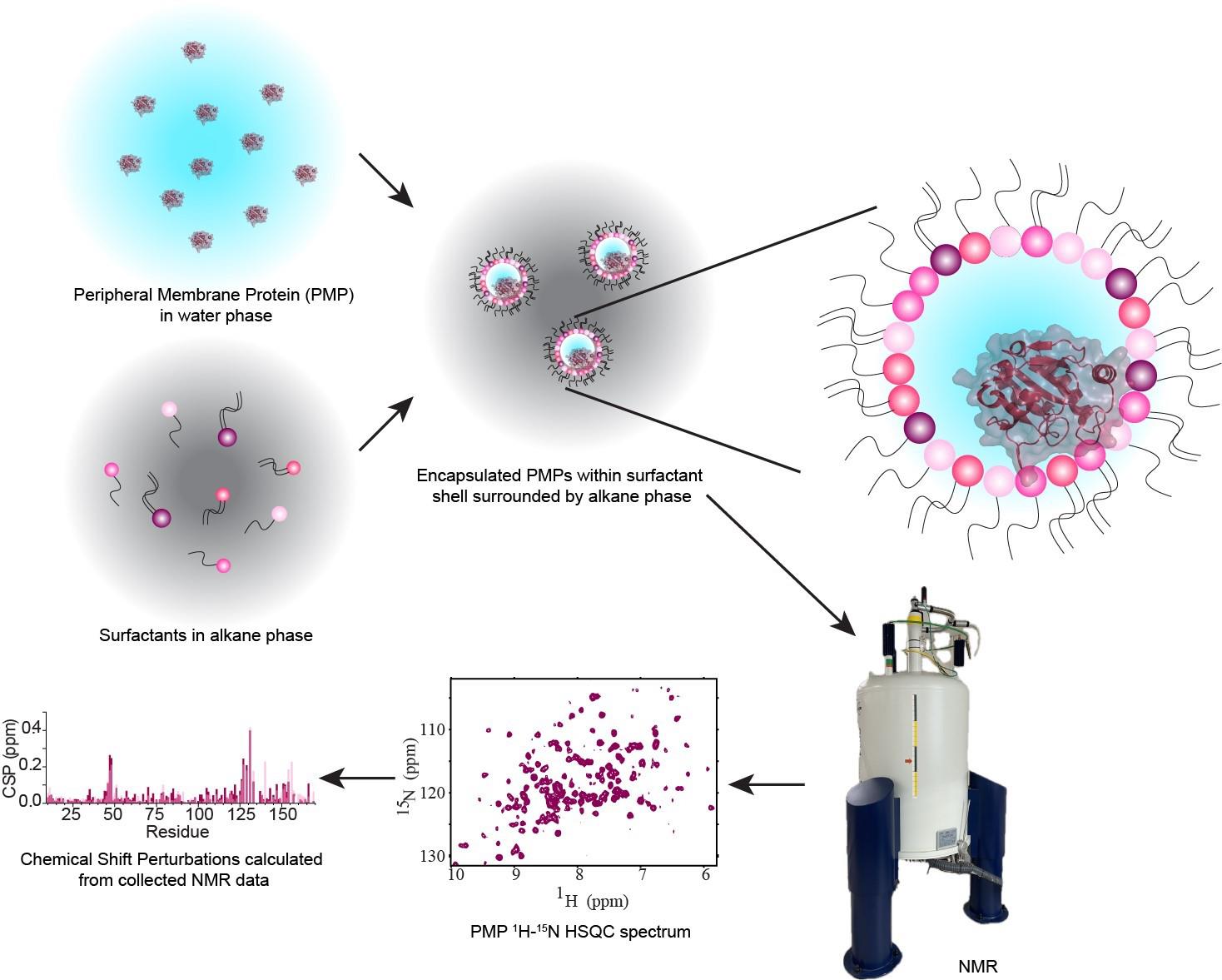
Background
Peripheral membrane proteins (PMPs) are water-soluble proteins that can bind, often reversibly, with membranes. Interactions between PMPs and their target membranes are of great interest due to the central role of these proteins in disease and biology. Currently, methods to accurately observe the protein–membrane interaction, especially at high resolution, are limited. Nuclear magnetic resonance (NMR) is well-suited to observe these interactions using micelles, bicelles, and nanodiscs. However, these membrane models are most commonly formulated from artificial components, limiting understanding in a biological context. Reverse micelles (RMs) have a long history in the study of proteins, including being used to house membrane-associated and membrane-integral proteins for NMR [1,2]. RMs house proteins within a nanoscale pool of water, surrounded by a hollow shell of surfactants with the hydrophilic headgroups interacting with the water and the hydrophobic tails pointing outward and serving to solubilize the RM in an alkane solvent. High-quality NMR spectra of proteins encapsulated within RMs are routinely attainable, enabled by the low-viscosity alkane solvent [3]. Recent developments of RMs have improved their properties as membrane mimetics through formulations using phosphocholine-based surfactants. Utilizing membrane-mimicking RMs (mmRMs), the PMP of interest can be observed interacting with the surfactant shell. This allows the mapping of PMP-membrane interaction surfaces and enables structural and functional studies using NMR. RMs formulated with naturally derived lipids represent an advancement in technology, allowing the study of PMPs and other proteins in a more native-like membrane mimetic [4]. The lipid extractions used for native reverse micelles (nRMs), including soy lecithin, bovine heart lipids, and porcine brain lipids, account for a wide range of lipid compositions. These compositions introduce a heterogeneous mixture of lipid headgroup and tail types, reflecting the complexity of biological membranes. Conversely, a formulation of RMs made entirely from phosphocholine-based surfactants can be used as a standard background to explore specific lipid interactions or for structural and functional studies that require a more homogeneous membrane model [5]. Previous applications of RMs to protein studies may also be extended to nRMs, including structural determination of PMPs [6], hydration dynamics measurements [7], experimental cosolvent mapping [8], and enhanced fragment screening [9]. Integral membrane proteins and proteins anchored through lipidations have previously been housed in RMs and would benefit from study in the more native-like environment described here [1,2]. The following protocol describes the construction of reverse micelles from lipids extracted from native sources or phosphocholine-based surfactants, for high-resolution NMR study of encapsulated PMPs in their membrane-bound state.
Materials and reagents
Biological materials
Protein of interest; in this protocol, glutathione peroxidase 4 (GPx4) is used as an example for encapsulation in reverse micelles. Protein was produced as previously described using recombinant 15N or 13C-15N isotopic labeling, expression, and purification from BL21(DE3) E. coli [10].
Reagents
Lecithin (VWR, catalog number: 10791-822)
Ethylenediaminetetraacetic acid (EDTA) (Fisher, catalog number: AAJ15694AE)
Sodium chloride (NaCl) (Fisher, catalog number: S271-1)
Bis-Tris (VWR, catalog number: 6976-37-0)
Tris base (Sigma-Aldrich, catalog number: 77-86-1)
Chloroform (Fisher, catalog number: 67-66-3)
Methanol (EM Science, catalog number: 67-56-1)
Distilled water
Hydrochloric acid (HCl) (Fisher, catalog number: SA49)
Dithiothreitol (DTT) (GoldBio, catalog number: DTT10)
Sodium sulfate (VWR, catalog number: 7757-82-6)
Brain total lipid extract (porcine) (PBL) (Avanti Polar Lipids, catalog number: 131101C)
Heart total extract (bovine) (BHL) (Avanti Polar Lipids, catalog number: 171201P)
Pentane (Alfa Aesar, catalog number: AA32449K2)
1-Hexanol (Sigma-Aldrich, catalog number: 111-27-3)
1,2-dilauroyl-sn-glycero-3-phosphocholine (DLPC) (Avanti Polar Lipids, catalog number: 850335C)
n-dodecylphosphocholine (DPC) (Avanti Polar Lipids, catalog number: 850336P)
D-pentane (Sigma-Aldrich, catalog number: 490482)
D2O (Sigma-Aldrich, catalog number: 7789-20-0)
Trimethyl phosphate (Sigma-Aldrich, catalog number: 512-56-1)
Bradford dye (Thermo Fisher, catalog number: 23238)
Solutions
5 M NaCl stock solution (see Recipes)
500 mM Bis-Tris pH 6.0 stock solution (see Recipes)
1 M DTT stock solution (see Recipes)
200 mM EDTA pH 6.0 stock solution (see Recipes)
6 M HCl stock solution (see Recipes)
Protein buffer (see Recipes)
Dilute protein buffer (see Recipes)
Mobile phase for soy lecithin chelation (see Recipes)
Recipes
5 M NaCl stock solution
*Note: Initially, add half of the distilled H2O to dissolve the NaCl; the rest of the volume should slowly be added as NaCl dissolves. The stock should be brought up to the final volume once all NaCl is dissolved to account for any volume displacement caused by the solid powder.
Reagent Final concentration Quantity or Volume NaCl 5 M 146.1 g H2O n/a *see Note Total n/a 500 mL 500 mM Bis-Tris pH 6.0 stock solution
*Note: Initially, add half of the distilled H2O to dissolve the Bis-Tris; most of the rest should slowly be added as Bis-Tris dissolves. Once the Bis-Tris is dissolved, add small increments of 6 M HCl into the solution to bring the pH to 6.0. The stock should be brought up to the final volume with distilled H2O once the Bis-Tris is pH-corrected to account for any volume displacement caused by the solid powder and HCl.
Reagent Final concentration Quantity or Volume Bis-Tris pH 6.0 500 mM 52.3 g H2O n/a *see Note pH corrected by HCl *see Note Total n/a 500 mL 1 M DTT stock solution
*Note: Initially, add half of the distilled H2O to dissolve the DTT; the rest should slowly be added as DTT dissolves. The stock should be brought up to the final volume once all DTT is dissolved to account for any volume displacement caused by the solid powder.
Reagent Final concentration Quantity or Volume DTT 1 M 1.54 g H2O n/a *see Note pH adjusted using HCl Total n/a 10 mL 200 mM EDTA pH 6.0 stock solution
*Note: Initially, add half of the distilled H2O to dissolve the EDTA; most of the rest should slowly be added as EDTA dissolves. Once the EDTA is dissolved, add small increments of 6 M HCl to the solution to bring the pH to 6.0. The stock should be brought up to the final volume with distilled H2O once the EDTA is pH-corrected to account for any volume displacement caused by the solid powder and HCl.
Reagent Final concentration Quantity or Volume EDTA 200 mM 2.92 g H2O n/a *see Note pH adjusted using HCl Total n/a 50 mL 6 M HCl stock solution
Reagent Final concentration Quantity or Volume HCl, 12 N 6 M 100 mL H2O n/a 100 mL Total 6 M 200 mL Protein buffer
Reagent Final concentration Quantity or Volume Bis-Tris pH 6.0, 500 mM stock 20 mM 2 mL NaCl, 5 M stock 100 mM 1 mL DTT, 1 M stock 20 mM 1 mL H2O n/a 46 mL Total n/a 50 mL Dilute protein buffer
Reagent Final concentration Quantity or Volume Bis-Tris pH 6.0, 500 mM stock 20 μM 2.4 μL NaCl, 5 M stock 100 μM 1.2 μL DTT, 1 M stock 20 μM 1.2 μL H2O n/a 59.952 mL Total n/a 60 mL Mobile phase for soy lecithin chelation
Note: Dry the solvents needed with sodium sulfate overnight and decant before the mobile phase is made.
Reagent Final concentration Quantity or Volume Chloroform n/a 10 mL Methanol n/a 5 mL Total n/a 15 mL
Laboratory supplies
1.5 mL glass vials with screw top lid (Sigma-Aldrich, catalog number: 854171)
Pierce Concentrator, PES membrane, 10K MWCO 0.5 mL (Thermo Fisher, catalog number: 88513)
Spin-XR UF 20 10K MWCO PES membrane 20 mL (Corning, catalog number: 431488)
Amicon Ultra-15 centrifugal filters regenerated cellulose membrane 3K 20 mL (Millipore Sigma, catalog number: UFC901024)
NMR tubes (New Era Enterprises, Inc., catalog number: NE-UL5-7)
1.5 mL microcentrifuge tubes (Cell treat, catalog number: 229441)
20 mL scintillation vials (Wheaton, catalog number: 03-341-25K)
Pipettes (Corning, catalog numbers: 4075, 4071, 4072, 4074)
Pipette tips (Fisherbrand/Olympus, catalog numbers: 02-717-134/22-119B, 24-150RL, 23-404)
Glass pipettes (Fisherbrand, catalog number: 13-678-6B)
Separation funnel (Chem Glass, catalog number: CG1743-09)
25 mL glass graduated cylinder (Chem Glass, catalog number: CG-8242-25)
Stir bar (Fisher Scientific, catalog number: 16-800-507)
Pipette bulbs (Fisherbrand, catalog number: 03-448-21)
Teflon tape (SPBel-Art, catalog number: 240200000)
Parafilm (Bemis Parafilm, catalog number: PM996)
Equipment
Pioneer analytical balance (Ohaus, model number: PX84/E)
Navigator analytical scale (Ohaus, model number: NV222)
Eppendorf centrifuge 5425 (Eppendorf, catalog number: 5405000646)
Eppendorf thermomixer (Eppendorf)
Note: The model used in this protocol was discontinued; a comparable model would be the Eppendorf Thermomixer, catalog number: 5384000020.
LSE digital dry bath (Corning, catalog number: 6875-SB)
Hereaus megafuge 8 centrifuge (Thermo Scientific, catalog number: 75007210)
ST plus series centrifuge (Sorvall, catalog number: 75009909)
Speed vacuum concentrator (Savant, catalog number: SVC100H)
nXDS vacuum pump (Edwards, catalog number: A73501983)
4 °C refrigerator (VWR, catalog number: 10819-904)
-80 °C freezer (Fisherbrand, catalog number: IUE30086FA)
Zetasizer nano (Malvern Panalytical, model: ZEN3600)
2800 ultrasonic bath (Branson, catalog number: M2800)
LSE vortex mixer (Corning, catalog number: 6778)
Mini centrifuge (Corning, catalog number: 6770)
AG centrifuge (Eppendorf, catalog number: 022620100)
Laboratory fume hood (Hamilton Laboratory Solutions, model: 61L)
Advance stir plate (VWR, catalog number: 76557-502)
Genesys 150 UV spectrophotometer (Thermo Scientific, catalog number: 840-300000)
pH meter (Oakton, catalog number: EW-35413-20)
High-field (≥ 500 MHz) NMR spectrometer, equipped with triple-resonance inverse probe
Software and datasets
Prism v10.2 (GraphPad, 02/5/2024)
NMRPipe (v11.4) [11]
NMRFAM-Sparky (v3.12, 04/15/2015)
Bruker Topspin (v4.1.3)
Procedure
文章信息
稿件历史记录
提交日期: Apr 16, 2024
接收日期: Jun 27, 2024
在线发布日期: Jul 11, 2024
出版日期: Jul 20, 2024
版权信息
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
如何引用
Walters, S. H. and Fuglestad, B. (2024). An NMR Approach for Investigating Membrane Protein–Lipid Interactions Using Native Reverse Micelles. Bio-protocol 14(14): e5039. DOI: 10.21769/BioProtoc.5039.
分类
生物物理学 > 核磁共振波谱法
生物化学 > 蛋白质 > 结构
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link