- EN - English
- CN - 中文
Electrophoretic Mobility Assay to Separate Supercoiled, Catenated, and Knotted DNA Molecules
电泳迁移率测定法用于分离超螺旋、连锁和打结的DNA分子
发布: 2024年05月05日第14卷第9期 DOI: 10.21769/BioProtoc.4983 浏览次数: 2138
评审: Pilar Villacampa AlcubierreAnonymous reviewer(s)
Abstract
Two-dimensional (2D) agarose gel electrophoresis is the method of choice to analyze DNA topology. The possibility to use E. coli strains with different genetic backgrounds in combination with nicking enzymes and different concentrations of norfloxacin improves the resolution of 2D gels to study the electrophoretic behavior of three different families of DNA topoisomers: supercoiled DNA molecules, post-replicative catenanes, and knotted DNA molecules. Here, we describe the materials and procedures required to optimize their separation by 2D gels. Understanding the differences in their electrophoretic behavior can help explain some important physical characteristics of these different types of DNA topoisomers.
Key features
• Preparative method to enrich DNA samples of supercoiled, catenated, and knotted families of topoisomers, later analyzed by 2D gels (or other techniques, e.g., microscopy).
• 2D gels facilitate the separation of the topoisomers of any given circular DNA molecule.
• Separation of DNA molecules with the same molecular masses but different shapes can be optimized by modifying the conditions of 2D gels.
• Evaluating the roles of electric field and agarose concentration on the electrophoretic mobility of DNA topoisomers sheds light on their physical characteristics.
Keywords: DNA topology (DNA拓扑)Graphical overview
Background
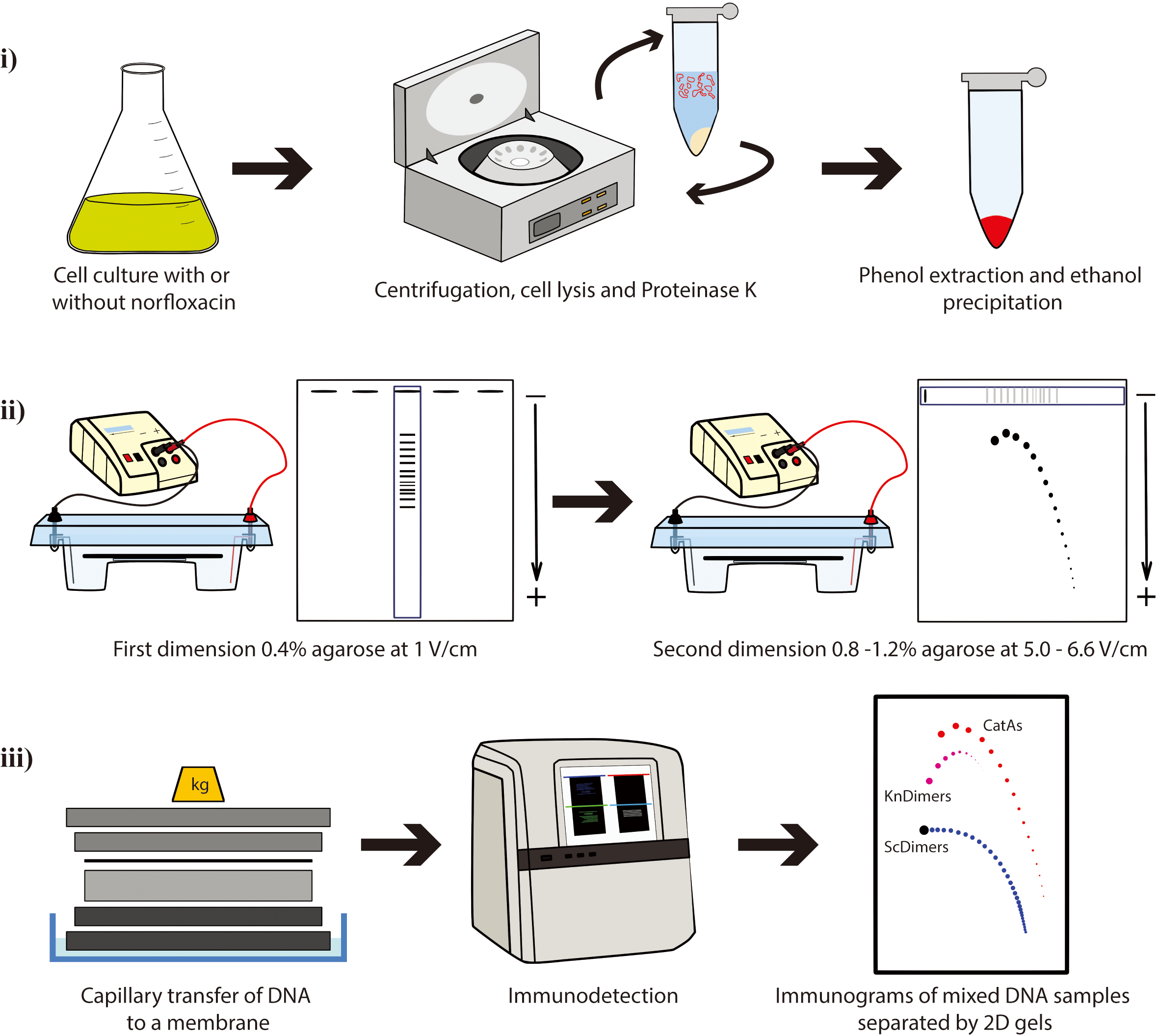
DNA molecules with different topology can be analyzed by the so-called single-molecule methods such as chromatin fiber autoradiography [1], dynamic molecular combing [2], transmission electron microscopy [3-5], atomic force microscopy [6], and magnetic tweezers [7,8]. DNA properties that are difficult to address experimentally can also be studied by computer simulations [9–13]. Two-dimensional (2D) agarose gel electrophoresis is the best experimental method currently available to allow the simultaneous identification of DNA molecules with different topology [e.g., supercoiled (Sc), catenated (Cats), and knotted (Kn) molecules]. This technique consists of two consecutive electrophoretic separations performed under different conditions and run at two orthogonal directions (4–8). The first dimension is resolved in a low-percentage (∼0.4%) agarose gel electrophoresis at a relatively low voltage (∼1 V/cm). The second dimension is run perpendicular to the first one so that an entire selected lane of the gel is used as replacement of the gel wells but in a higher-percentage (∼1%) agarose gel electrophoresis at higher electric field (∼5–6.6 V/cm). 2D gels were originally designed by Bell and Byers to separate branched and linear molecules [14], and it was early noticed that this method could also be successfully applied to study DNA topology. 2D gels were adapted to examine simultaneously thousands of molecules with different DNA topology, such as Sc forms, Kn forms, partially replicated forms (named pre-catenanes) with or without reversed forks, fully replicated catenanes (Cats), and replication intermediates (RIs) containing knotted bubbles [4,6,15–28]. 2D agarose gel electrophoresis has been extensively used to investigate the activity of topoisomerases in vitro and in vivo [29,30]. Additionally, 2D gels can also be used as a preparative method to enrich samples for specific DNA molecules that can be later examined by different techniques [4,6,18,19,31,32].
Plasmids are invaluable tools as a model to study DNA topology. The advantages of plasmids include their ease of isolation and the ability to quantitatively measure DNA supercoiling, knotting, and catenation in purified DNA samples [33]. Here, we present a protocol where 2D gels are used to analyze electrophoretic mobility of three families of topoisomers with the same molecular mass (8,766 bp): supercoiled dimers (ScDimers), monomeric-nicked catenanes (CatAs), and nicked-knotted dimers (KnDimers). We use monomeric (4,383 bp) or dimeric forms (8,766 bp) of pBR18, a derivative of pBR322 where the tetracycline resistance promoter was replaced with the poly-linker of pUC18 [34] to transform three different Escherichia coli strains. We describe the steps required to:
i. Obtain the three families of DNA samples for the analysis by 2D gels;
ii. Optimize their separation by 2D gels based on their extent of supercoiling, catenation, and knotting;
iii. Visualize them by non-radioactive detection.
Materials and reagents
Biological materials
Escherichia coli strains
DH5αF’: F’/gyrA96(Nalr) recA1 relA1 endA1 thi-1 hsdR17 (rk-mk+) glnV44 deoR Δ (lacZYA argF) U169[F80dΔ(lacZ)M15] (kindly gifted by Santiago Rodríguez de Córdoba) [35]
parE10: W3110 F- except [parE10 recA] (kindly gifted by Ian Grainge) [36]
LZ38 F-λ (P80 red114 xis-l cl857) zei-723::Tn10 parCK84::kanR (kindly gifted by Lynn Zechiedrich) [37,38]
Plasmid:
pBR18 (4,383 pb): a derivative of pBR322 where the tetracycline resistance promoter between EcoRI and HindIII was replaced with the poly-linker of pUC18 [32]
Luria-Broth (LB) (Invitrogen, catalog number: 12795084)
Ethanol absolute, molecular biology grade (Sigma, catalog number: 64-17-5)
Agarose (Seakem LE, Lonza, catalog number: 50004)
Norfloxacin (Abcam, catalog number: 70458-96-7)
Ethidium bromide (Sigma, catalog number: 1239-45-8)
Digoxigenin-High Prime kit (Roche, catalog number: 11585614910)
Anti-Digoxigenin-AP conjugate antibody (Roche, catalog number: 11093274910)
CDP-Star (Perkin Elmer, catalog number: NEL601001KT)
Brij® 58 (Sigma-Aldrich, catalog number: P-5884)
Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750)
Tris-HCl (Sigma, catalog number: T2913)
Ethylenedinitrilotetraacetic acid, disodium salt (EDTA) (Sigma, catalog number: E7889)
Tris base (Sigma, catalog number: T1503)
Boric acid (Sigma, catalog number: PHR2664)
Hydrochloric acid (Sigma-Aldrich, catalog number: 258148)
Proteinase K, recombinant (Roche, catalog number: 3115879001)
Nb.BsmI 10,000 U/mL (New England Biolabs, catalog number: R0706S)
Calcium chloride (Flinn Scientific, catalog number: C0234)
Lambda DNA/HindIII Marker (Invitrogen, catalog number: SM0101)
BlueJuiceTM gel loading dye (10×) (Thermo Scientific, catalog number: 10816015)
Magnesium sulfate (MP Biomedicals, catalog number: 0219483394)
Sodium chloride (MP Biomedicals, catalog number: 0215257591)
Sucrose (Thermo Scientific Chemicals, catalog number: A15583.36)
Lysozyme (Thermo Scientific, catalog number: 10249843)
Polyethylene glycol (PEG) 6000 (Thermo Scientific, catalog number: J19972.A1)
Sodium hydroxide (Fisher Chemical, catalog number: 10675692)
Tri-sodium citrate (Fisher Chemical, catalog number: 10112880)
Disodium phosphate (MP Biomedicals, catalog number: 02191440.5)
Dextran sulfate (MP Biomedicals, catalog number: 11417880)
Sodium azide (Sigma, catalog number: S2002)
Sodium dodecyl sulfate (SDS) (Sigma, catalog number: 151-21-3)
Blotto (regular powdered milk at supermarket/pharmacy)
Sonicated and denatured salmon sperm DNA (Invitrogen, catalog number: 10605543)
Phenol:chloroform:isoamyl alcohol (25:24:1) equilibrated with 10 mM Tris–HCl, pH 8.0. (Sigma, catalog number: P3803)
0.5 M EDTA, pH 8.0
For bacterial transformation (Section A):
1 M magnesium sulfate
0.1 M calcium chloride
For isolation of plasmid DNA (Section B):
Tris-EDTA (TE) (see Recipes)
Sodium chloride-Tris-EDTA (STE) (see Recipes)
25% sucrose (see Recipes)
Lysozyme–RNase A solution (see Recipes)
Lysis buffer (see Recipes)
25% PEG 6000 in TE (see Recipes)
Proteinase K buffer (20 mg/mL) (see Recipes)
For 2D agarose gel electrophoresis (Section E):
10× Tris Borate EDTA (TBE) (see Recipes)
For Southern blotting (Section F):
Depurination solution (see Recipes)
Transfer solution (see Recipes)
20× saline sodium citrate (SSC) (see Recipes)
For nonradioactive hybridization (Section G):
20× saline sodium phosphate EDTA (SSPE) (see Recipes)
20% dextran sulfate (see Recipes)
20% SDS (see Recipes)
Prehybridization/hybridization solution (see Recipes)
Recipes
For isolation of plasmid DNA (Section B):
Tris-EDTA (TE)
10 mM Tris-HCl
1 mM EDTA, pH 8.0
Ultrapure water
Note: Sterilize and store at room temperature. Stable for one year.
Sodium chloride-Tris-EDTA (STE)
0.1 M sodium chloride
10 mM Tris-HCl, pH 8.0
1 mM EDTA, pH 8.0
Ultrapure water
Note: Sterilize and store at 4 °C. Stable for one year.
25% Sucrose
25% sucrose
0.25 M Tris-HCl, pH 8.0
Note: Sterilize and store at 4 °C. Stable for one month.
Lysozyme–RNase A solution
10 mg/mL lysozyme
0.2 mg/mL RNase A
0.25 M Tris-HCl, pH 8.0.
0.25 M EDTA, pH 8.0
Note: Prepare immediately prior to use; mix by inverting the tube and keep on ice.
Lysis buffer
50 mM Tris–HCl, pH 8.0
63 mM EDTA
1% Brij® 58
0.4% sodium deoxycholate
Ultrapure water
Note: Difficult to dissolve, use a magnetic stirrer. Stable for six months.
25% PEG 6000 in TE
25% PEG 6000
1.25 M sodium chloride
Note: Difficult to dissolve, use a magnetic stirrer. Store at 4 °C. Stable for two years.
Proteinase K buffer (20 mg/mL)
0.1 M sodium chloride
10 mM Tris-HCl, pH 8.0
1 mM EDTA, pH 8.0
0.1% SDS
Ultrapure water
Note: Preheat at 37 °C and add 100 μg/mL of Proteinase K before use. Proteinase K buffer should be stored at -20 °C for up to two years to maintain its activity.
For 2D agarose gel electrophoresis (Section E):
10× Tris Borate EDTA (TBE)
1.00 M Tris base
1.00 M boric acid
0.02 M EDTA
Ultrapure water
pH 8.2–8.4
Note: Prepare a 1:10 dilution with ultrapure water to make 1× TBE. Store at room temperature. Stable for one year.
For Southern blotting (Section F):
Depurination solution
0.25 M hydrochloric acid
Ultrapure water
Note: Prepare immediately prior to use.
Transfer solution
0.4 M sodium hydroxide
Ultrapure water
Note: Prepare immediately prior to use.
20× saline sodium citrate (SSC)
3 M sodium chloride
0.3 M Tri-sodium citrate
Ultrapure water
Note: Store at room temperature. Stable for two years.
For nonradioactive hybridization (Section G):
20× saline sodium phosphate EDTA (SSPE)
3.6 M sodium chloride
0.2 M disodium phosphate
20 mM EDTA
Ultrapure water
Note: Store at room temperature. Stable for two years.
20% dextran sulfate
20 g of dextran sulfate
0.2 g of sodium azide
Ultrapure water
Note: Prepare aliquots and freeze (-20 °C). Stock solutions are stable for up to six months.
20% SDS
3 M sodium chloride
0.3 M sodium citrate
Ultrapure water
pH 7.0
Note: Store at room temperature. Stable for one year.
Prehybridization/hybridization solution
2× SSPE
1% SDS
0.5% Blotto
10% dextran sulfate
0.5 mg/mL sonicated and denatured salmon sperm DNA
Ultrapure water
Note: Prepare aliquots and freeze (-20 °C). Stock solutions are stable for up to 2 years.
Laboratory supplies
500 mL centrifuge bottles (Thermo Scientific, catalog number: 010-1493)
50 mL high-speed centrifuge tubes (Thermo Scientific, catalog number: 3118-0085)
50 mL Falcon tubes (Fisherbrand, catalog number: 11512303)
15 mL Falcon tubes (Fisherbrand, catalog number: 11765075)
1.5 mL microcentrifuge tubes (Thermo Scientific, PierceTM, catalog number: 10177443)
Whatman filter paper (GE Healthcare, catalog number: 3030917)
Zeta-Probe membrane (Bio-Rad, catalog number: 1620159)
Glass or plastic pipette
Film (Fuji, catalog number: 4741019236)
Cell spreader (Sigma-Aldrich, model: Cell Spreader silver stainless steel, bar L 33 mm, catalog number: HS86655)
Equipment
Superspeed floor centrifuge (Thermo Scientific, catalog number: 75006580)
Fixed-angle rotor for 500 mL bottles (Thermo Scientific, catalog number: 096-062375)
Swinging-bucket rotor for 50 mL round or conical tubes (Thermo Scientific, catalog number: 75003010)
Horizontal electrophoresis system with inter-electrode distances of at least 30 cm that can hold a 15 × 20 cm tray (Bio-Rad, catalog number: 1704403)
Basic power supply (Bio-Rad, catalog number: 11645050)
UV transilluminator (Biorad, model: Gel Doc XR+ System, catalog number: 1708195)
Microbiological shaking incubator (VWR, catalog number: 76628-472)
Thermoblock (Labnet, model: AccuBlock, catalog number: D1301)
Vacuum pump (Millipore, model: Millivac-Mini Vacuum Pump XF5423050)
Hybridization oven (Techne®, Hybrigene, catalog number: 445-0024)
X-ray film cassette or ChemiDocTM Imaging System (Bio-Rad, catalog number: 12003153)
Microcentrifuge (Eppendorf, model: 5418R, catalog number: EPP5401000013)
Software and datasets
ImageJ (https://imagej.nih.gov/ij/download.html) (Access date, 02/01/2024)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Cebrián, J., Martínez, V., Hernández, P., Krimer, D. B., Martínez-Robles, M. L., Schvartzman, J. B. and Fernández-Nestosa, M. J. (2024). Electrophoretic Mobility Assay to Separate Supercoiled, Catenated, and Knotted DNA Molecules. Bio-protocol 14(9): e4983. DOI: 10.21769/BioProtoc.4983.
分类
分子生物学 > DNA > DNA 结构
分子生物学 > DNA > 电泳
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link