- EN - English
- CN - 中文
CRISPR/dCas9-Tet1-Mediated DNA Methylation Editing
CRISPR/dCas9-Tet1介导的DNA甲基化编辑
发布: 2024年04月20日第14卷第8期 DOI: 10.21769/BioProtoc.4976 浏览次数: 4110
评审: Clara Morral MartinezPhilipp WörsdörferAnonymous reviewer(s)
Abstract
DNA methylation is a key epigenetic mechanism underlying many biological processes, and its aberrant regulation has been tightly associated with various human diseases. Precise manipulation of DNA methylation holds the promise to advance our understanding of this critical mechanism and to develop novel therapeutic methods. Previously, we were only able to alter genome-wide DNA methylation by treating with small molecules (e.g., 5-Aza-2-deoxycytidine) or perturbing relevant genes (e.g., DNA methyltransferase) targetlessly, which makes it challenging to investigate the functional significance of this epigenetic mark at specific genomic loci. By fusing the catalytic domain of a key enzyme in the DNA demethylation process (Ten-eleven translocation dioxygenases 1, Tet1) with a reprogrammable sequence-specific DNA-targeting molecular protein, dCas9, we developed a DNA methylation editing tool (dCas9-Tet1) to demethylate specific genomic loci in a targeted manner. This dCas9-Tet1 system allows us to study the role of DNA methylation at almost any given loci with only the replacement of a single-guide RNA. Here, we describe a protocol that enables modular and scalable manipulation of DNA methylation at specific genomic loci in various cell cultures with high efficiency and specificity using the dCas9-Tet1 system.
Key features
• Precisely editing the DNA methylation of specific genomic loci in a targeted manner.
• Fine-tuning gene expression without changing DNA sequence.
• Applicable to many types of cell cultures and with the potential for ex vitro and in vivo applications.
Keywords: CRISPR (CRISPR)Graphical overview
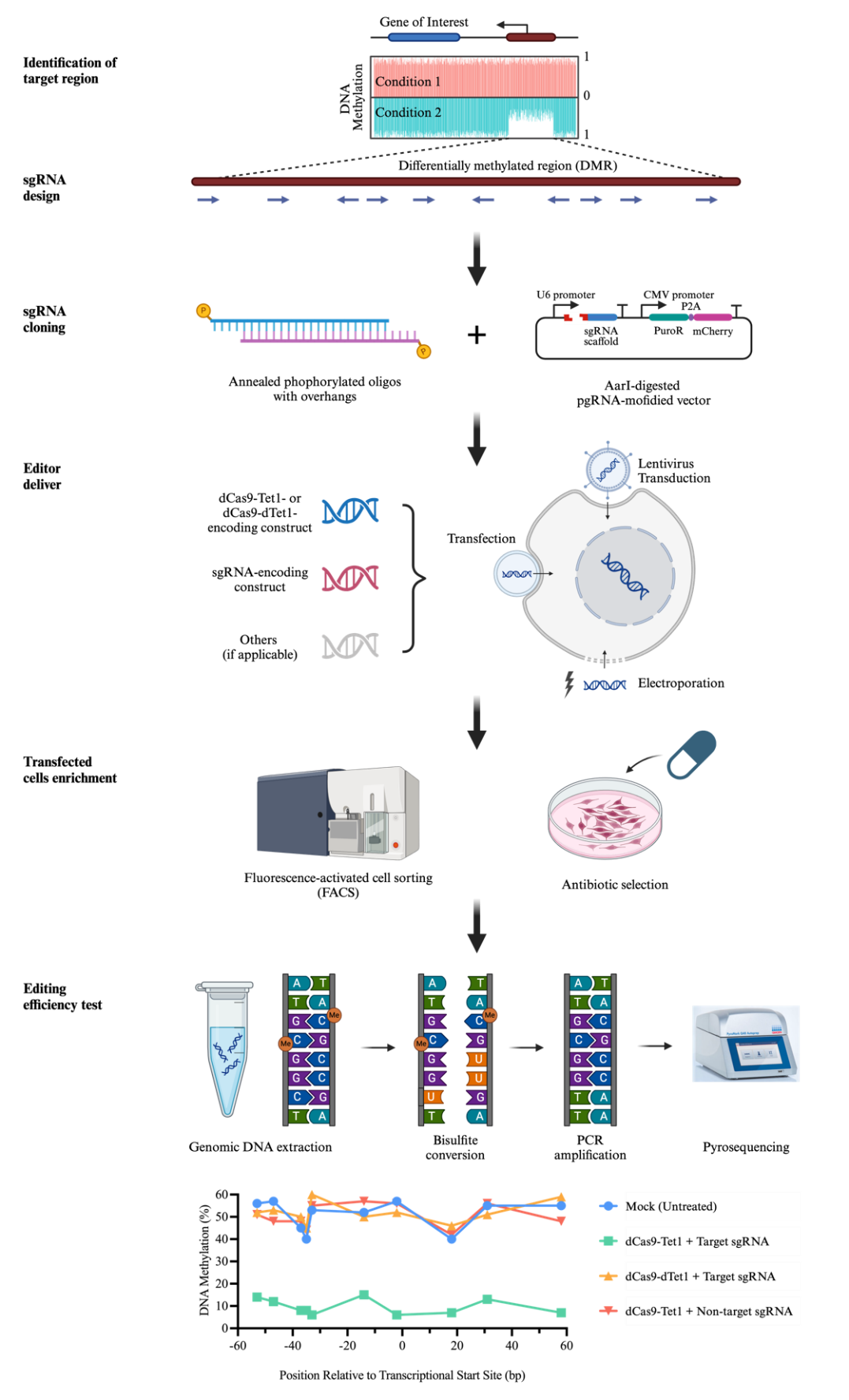
Overview of dCas9-Tet1-mediated DNA methylation editing
Background
DNA methylation is one of the most important epigenetic modifications, which refers to the covalent modification of the cytosine of DNA by a methyl group. In mammals, DNA methylation often occurs in the context of cytosine-phosphate-guanosine (CpG) residues [1]. It plays a critical role in various biological processes including gene regulation, genomic imprinting, and X chromosome inactivation, as well as the preservation of chromosome stability [2,3]. The proper regulation of DNA methylation is pivotal in maintaining normal cellular activities, and its dysregulation has been tightly associated with a myriad of human diseases [4].
Given its significance, the ability to manipulate DNA methylation with precision could revolutionize both basic research and therapeutic fields. Over the years, several epigenetic editing tools have been developed to manipulate DNA methylation at specific sites by tethering DNA methylation-associated effectors to sequence-specific DNA-binding domains, comprising mainly zinc finger proteins (ZFPs), transcription activator-like effectors (TALEs), and the clustered regularly interspaced short palindromic repeats (CRISPR) system [5–7]. Our lab pioneered a CRISPR-based DNA-methylation editing system using the fusion of a catalytically dead Cas9 (dCas9) with Ten-eleven translocation 1 (Tet1) hydroxylase catalytic domain (dCas9-Tet1), allowing for the erasure of DNA methylation in the mammalian genome with high specificity [8]. It has been proved as a powerful technology in many aspects, including mechanistically dissecting the functional significance of DNA methylation as well as treating incurable diseases by rewriting the DNA methylation of disease-causing genes. For instance, we demonstrated that targeted demethylation of the MyoD distal enhancer by dCas9-Tet1 along with 5-Aza (5-Aza-2-deoxycytidine, a DNA methylation inhibitor) treatment facilitated myogenic reprogramming of fibroblasts [8]. With the application of dCas9-Tet1, we also studied the hypermethylation of the CGG repeat expansion mutation at the 5’ UTR of fragile X mental retardation 1 (FMR1) gene. We demonstrated that demethylation of the CGG repeats unlocked the epigenetic silencing of FMR1 and restored FMRP expression in Fragile X syndrome cells [9]. More recently, we reported that dCas9-Tet1-mediated DNA demethylation of the promoter of methyl CpG-binding protein 2 (MECP2) gene can reactivate the expression of MECP2 from the inactive X chromosome in Rett syndrome (RTT)-like human embryonic stem cells (hESCs) and in hESCs-derived neurons, which could be a potential therapeutic approach for Rett syndrome [10].
Compared to ZFPs- or TALEs-based approaches, our methods displayed higher efficacy, specificity, and resolution of DNA methylation editing [8]. Importantly, unlike ZFPs and TALEs, which require design and optimization of corresponding distinct proteins to target different DNA sequences, only a replacement of single-guide RNA (sgRNA) is needed for CRISPR/dCas9 system to target different genomic loci. The versatility to flexibly target almost any given genomic loci enable researchers to precisely manipulate DNA methylation with ease. Nevertheless, the size and structural complexity of dCas9-Tet1 system might limit its application in vivo.
Herein, we describe a step-by-step protocol for editing DNA methylation in cell cultures using the dCas9-Tet1 system. This protocol is divided into three subsections to outline 1) the identification of sgRNA target sequences and clone sequences into sgRNA scaffold construct, 2) the delivery of the constructs encoding dCas9-Tet1 and sgRNA into the cells of interest, and 3) the examination of editing results by pyrosequencing.
Materials and reagents
Biological materials
Human embryonic kidney (HEK) 293T cells (ATCC, catalog number: CRL-11268)
Human embryonic stem cells (hESCs) [National Institutes of Health (NIH) registration number: WIBR-2, #29]
Mouse embryonic fibroblasts (MEFs) (ATCC NIH3T3, catalog number: CRL-1658 or derived from other labs)
Stbl3 chemical competent cells (Invitrogen, catalog number: C737303)
pgRNA-modified (Addgene, catalog number: 84477)
Fuw-dCas9-Tet1-P2A-BFP (Addgene, catalog number: 108245)
Fuw-dCas9-dTet1-P2A-BFP (Addgene, catalog number: 108246)
PiggyBac transposase (Lab stock, available upon request)
dCas9-Tet1 on PiggyBac transposon vector (Lab stock, available upon request)
pCMV-dR8.74 (Addgene, catalog number: 22036)
pCMV-VSVG (Addgene, catalog number: 8454)
Reagents
DNA oligos (Eton Bioscience, customized order)
T4 ligase and its buffer (New England Biolabs, catalog number: M0202)
T4 polynucleotide kinase (PNK) (New England Biolabs, catalog number: M0201)
AarI enzyme (Thermo Scientific, catalog number: ER1581)
Agarose (Fisher Scientific, catalog number: BP1356-500)
Zymoclean Gel DNA Recovery kit (Zymo Research, catalog number: D4007)
S.O.C medium (Invitrogen, catalog number: 15544034)
mTeSR1 medium (Stemcell, catalog number: 85850)
LB Broth, Miller (Fisher Scientific, catalog number: BP1426-500)
Agar (Fisher Scientific, catalog number: BP1423-500)
Carbenicillin disodium salt (Sigma Aldrich, catalog number: C1389)
E.Z.N.A. plasmid mini kit I (Omega Biotek Inc, catalog number: D6943)
E.Z.N.A. plasmid mini kit II (Omega Biotek Inc, catalog number: D6945)
ZymoPURE II plasmid purification kit, Maxiprep (Zymo Research, catalog number: D4203)
X-tremeGENE DNA transfection reagent (Sigma Aldrich, catalog number: 06365787001)
Opti-MEM I reduced serum medium (Gibco, catalog number: 11058021)
DMEM, high glucose (Gibco, catalog number: 11095092)
Fetal bovine serum (FBS) (Gibco, catalog number: 10082-147)
200 mM L-glutamine (Gibco, catalog number: 25030081)
MEM non-essential amino acids solution, 100× (Gibco, catalog number: 11140076)
Penicillin-Streptomycin (10,000 U/mL) (Gibco, catalog number: 15140122)
Trypsin-EDTA (0.25%), phenol red (Gibco, catalog number: 25200056)
DMEM/F12 medium (Invitrogen, catalog number: 11320-033)
Knockout serum replacement (Invitrogen, catalog number: 10828028)
β-mercaptoethanol (Life Tech, catalog number: 21985023)
Fibroblast growth factor 2 (FGF2) (Gibco, catalog number: PHG0263)
Calcium- and magnesium-free phosphate-buffered saline (PBS) (Gibco, catalog number: 10010023)
Bovine serum albumin (BSA) Fraction V solution (7.5%) (Gibco, catalog number: 15260037)
0.5 M EDTA, pH 8.0 (Invitrogen, catalog number: 15575038)
1 M HEPES (Hyclone, catalog number: SH30237.01)
DNeasy blood & tissue kit (Qiagen, catalog number: 69504)
QIAamp DNA micro kit (Qiagen, catalog number: 56304)
EZ DNA Methylation-Gold kit (Zymo Research, catalog number: D5006)
PyroMark PCR Master Mix kit (Qiagen, catalog number: 978703)
PyroMark Q48 Advanced CpG Reagents (4 × 48) (Qiagen, catalog number: 974002)
Rock inhibitor (Sigma-Aldrich, catalog number: S4317)
Doxycycline (Sigma-Aldrich, catalog number: D9891)
Solutions
HEK293T cell culture medium (see Recipes)
hESCs culture medium (see Recipes)
FACS buffer (see Recipes)
Recipes
HEK293T cell culture medium
Reagent Final concentration Volume DMEM medium n/a 435 mL FBS 10% 50 mL 200 mM L-glutamine 2 mM 5 mL Non-essential amino acids 1% 5 mL Penicillin-streptomycin 1% (100 U/mL) 5 mL Total n/a 500 mL hESCs culture medium
Reagent Final concentration Quantity or Volume DMEM/F12 medium n/a 385 mL FBS 15% 75 mL Knockout serum replacement 5% 25 mL 200 mM L-glutamine 2 mM 5 mL Non-essential amino acids 1% 5 mL Penicillin-streptomycin 1% (100 U/mL) 5 mL β-mercaptoethanol 0.1 mM 0.5 mL FGF2 4 ng/mL 2 μg Total n/a 500 mL FACS buffer
Reagent Final concentration Volume PBS n/a 142.62 mL 7.5% BSA 0.5% 5 mL 0.5 M EDTA 0.5 mM 150 μL 1 M HEPES 15 mM 2.25 mL Total n/a 150 mL
Laboratory supplies
1.5 mL microcentrifuge tube (Axygen, catalog number: MCT-175-C)
PCR tube (Axygen, catalog number: PCR-0208-CP-C)
12-well plate (Corning, catalog number: 3513)
96-well plate (Corning, catalog number: 3596)
T-175 flask (Corning, catalog number: 431080)
0.22 μm filter (Corning, catalog number: 430513)
0.45 μm low protein binding filter (Thermo Scientific, catalog number: 121-0045)
Ultracentrifuge tube (Beckman Coulter, catalog number: 344058)
0.4 cm electroporation cuvette (Bio-Rad, catalog number: 1652081)
Equipment
Thermocycler (Applied Biosystem, ProFlex)
PyroMark Q48 Autoprep sequencer (Qiagen, catalog number: 9002471)
Bio-Rad gene pulser Xcell electroporation systems (Bio-Rad, catalog number: 1652660)
FACSAria cell sorter (BD, model: BD FACSAria™ II)
Fluorescence microscope (Nikon, model: Eclipse TS2R)
Centrifuge (Qiagen, model: 5810 with rotor A-2-DWP-AT)
Ultracentrifuge and rotor (Beckman, model: SW32Ti)
Software and datasets
PyroMark Assay Design (Version 2.0.2, 1/10/2024)
PyroMark Q48 Autoprep (Version 4.3.3, 1/10/2024)
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Qian, J. and Liu, S. X. (2024). CRISPR/dCas9-Tet1-Mediated DNA Methylation Editing. Bio-protocol 14(8): e4976. DOI: 10.21769/BioProtoc.4976.
- Qian, J., Guan, X., Xie, B., Xu, C., Niu, J., Tang, X., Li, C. H., Colecraft, H. M., Jaenisch, R. and Liu, X. S. (2023). Multiplex epigenome editing of MECP2 to rescue Rett syndrome neurons. Sci. Transl. Med. 15(679): eadd4666. https://doi.org/10.1126/scitranslmed.add4666.
分类
分子生物学 > DNA > DNA 修饰
生物科学 > 生物技术 > CRISPR/Cas9
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link