- EN - English
- CN - 中文
An Optimized P. berghei Liver Stage–HepG2 Infection Model for Simultaneous Quantitative Bioimaging of Host and Parasite Nascent Proteomes
用于宿主和寄生虫新生蛋白质组同时定量生物成像的优化伯氏疟原虫肝期- HepG2感染模型
发布: 2024年03月05日第14卷第5期 DOI: 10.21769/BioProtoc.4952 浏览次数: 1808
评审: Clara Morral MartinezRaniki KumariAnonymous reviewer(s)

相关实验方案
使用 Seahorse XFe96 细胞外通量分析仪实时分析弓形虫寄生虫的线粒体电子传递链功能
Jenni A. Hayward [...] Giel G. van Dooren
2022年01月05日 4025 阅读
Abstract
The Plasmodium parasites that cause malaria undergo an obligate, asymptomatic developmental stage in the host liver before initiating the symptomatic blood-stage infection. The parasite liver stage is a key intervention point for antimalarial chemoprophylaxis: successful targeting of liver-stage parasites prevents disease development in individuals and can help to reduce parasite transmission in populations, as the gametocyte forms that transmit infection to mosquitos are exclusively found in the blood stage. Antimalarial drugs that can target multiple parasite stages are thus highly desirable, and one emerging cellular target for such multistage active compounds is the process of protein synthesis or translation. Quantitative study of liver stage translation, and thus mechanistic evaluation of translation inhibitors against liver stage parasites, is not amenable to the methods allowing quantification of asexual blood stage translation, such as radiolabeled amino acid incorporation or lysate-based translation of reporter transcripts. Here, we present a method using o-propargyl puromycin (OPP) labeling of host and parasite nascent proteomes in the P. berghei-HepG2 infection model, followed by automated confocal image acquisition and computational separation of P. berghei vs. H. sapiens nascent proteome signals to allow simultaneous readout of the effects of translation inhibitors on both host and parasite. This protocol details our HepG2 cell culture and infected monolayer handling optimized for microscopy, our OPP labeling workflow, and our approach to automated confocal imaging, image processing, and data analysis.
Key features
• Uses the o-propargyl puromycin labeling technique developed by Liu et al. to quantitatively analyze protein synthesis in Plasmodium berghei liver-stage parasites in actively translating hepatoma cells.
• This quantitative approach should be adaptable for other puromycin-sensitive intracellular pathogens residing in actively translating host cells.
• The P. berghei–infected HepG2 recovery and reseeding protocol presented here is of use in applications beyond nascent proteome labeling and quantification.
Graphical overview
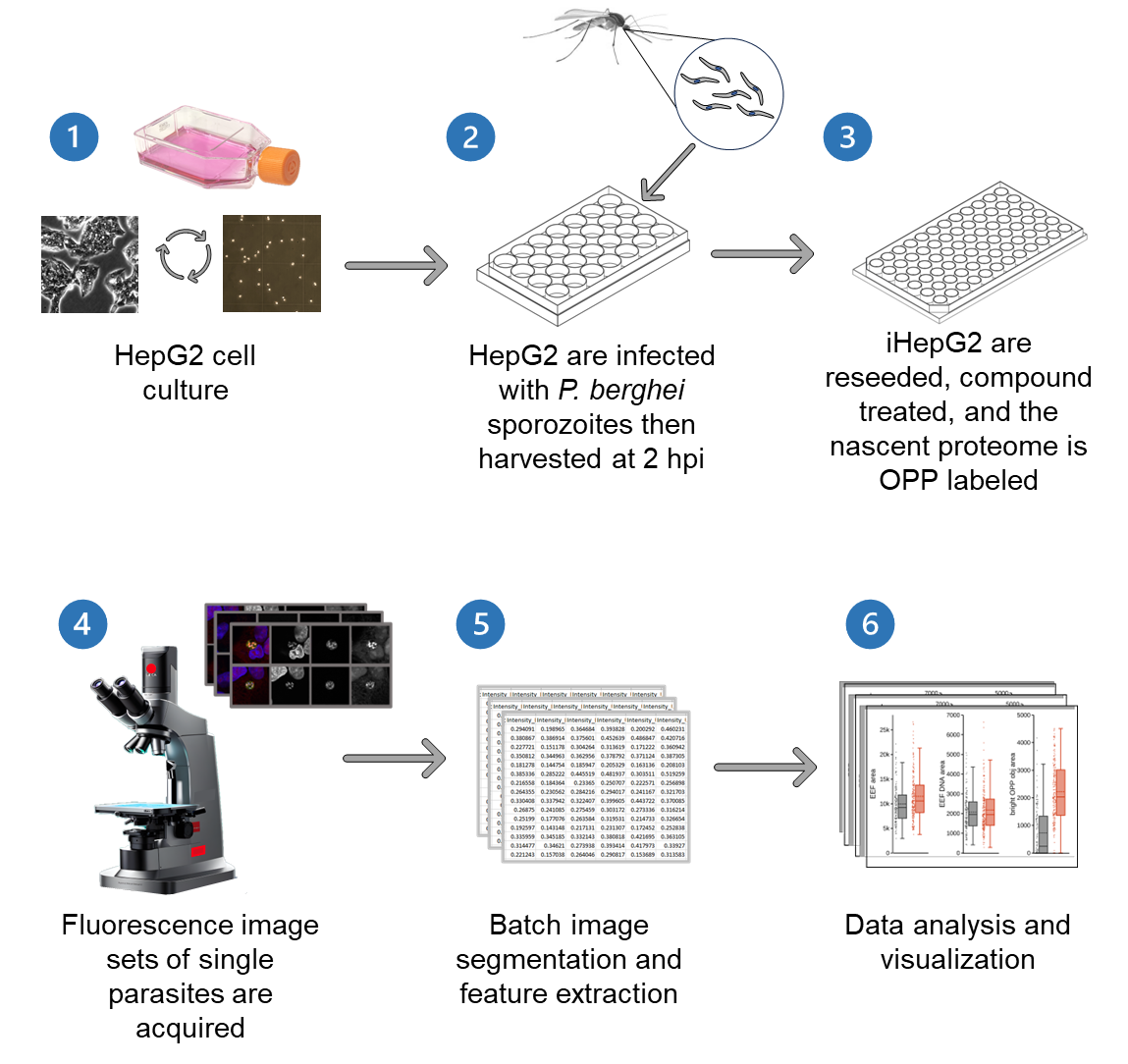
Background
Plasmodium parasites, the causative agents of malaria, have a complex lifecycle spanning both mosquito and human hosts. The first obligate step for parasite development in humans occurs in the liver following the introduction of motile parasites forms, called sporozoites, via an anopheline mosquito bite. Here, the liver-stage parasites, or exoerythrocytic forms (EEFs), undergo an extensive growth and replication process entirely within a parasitophorous vacuole in the cytoplasm of a hepatocyte; a single sporozoite will result in the formation of thousands of hepatic merozoites, which initiate the blood stage of infection [1]. While the continuous asexual replication cycles of blood-stage parasites can give rise to the signs and symptoms of malaria, the liver stage is asymptomatic and a key target for both vaccination strategies and chemoprophylaxis. The most widely used model species for studying the Plasmodium liver stage is P. berghei, a rodent malaria parasite that is capable of invading and growing in a variety of cell lines, including human hepatoma cell lines, during its liver-stage development [2,3]. Several immortalized cell lines are in regular use today, including HepG2 (human hepatoma), Huh-7 (human hepatoma), and HeLa (human cervical adenocarcinoma). Hepatocytes are highly specialized, polarized epithelial cells making up the liver parenchyma, and HepG2 are unique amongst the cell lines used for P. berghei liver-stage infections in their substantial maintenance of hepatic polarity, thus providing the closest in vitro cellular environment to that of the native host hepatocytes within a mammalian liver [4,5]. Hepatocytes in the liver are arranged in chords within hexagonal lobules in an architecture where each hepatocyte has direct basal surface contact with blood flowing through the fenestrated sinusoidal endothelium, while its multiple apical surfaces form the lumina into which bile is secreted at lateral points of contact with other hepatocytes [4,5]. The regenerative capacity of the liver relies on the ability of quiescent hepatocytes to reenter the cell cycle and divide, and the organizing principle that keeps the architecture of the chords intact relies on precise alignment of the daughter cells along the sinusoid [6]. These structural and functional particularities of the native hepatocyte environment have consequences for the in vitro growth of polarized HepG2 cells, which have a reputation for forming 3-dimensional layers or clumps in the absence of the liver architecture, which can hinder quantitative bioimaging in the P. berghei–HepG2 infection model. Careful attention to HepG2 culture and passage conditions can minimize this, allowing quantitative bioimaging of both host and parasite characteristics like translational output, which we present here.
Core cellular processes are somewhat difficult to study in Plasmodium liver stages. Protocols for routine isolation of EEFs have not been reported, so signals from the parasite must be separated from those of both infected and uninfected hepatocytes in the culture. While quantitative translation assays exist for Plasmodium blood-stage parasites [7–9], to our knowledge this is the first protocol for direct quantification of liver-stage translation in single cells and populations in the P. berghei–HepG2 infection model [10,11]. This assay relies on the use of the modified puromycin analogue, o-propargyl puromycin (OPP) [12], to label the HepG2 and P. berghei nascent proteomes during a 30 min window, followed by fixation, click chemistry–based attachment of a fluorophore to the OPP-labeled nascent polypeptides, and immunolabeling of the parasites, followed by confocal imaging, batch image segmentation and feature extraction to computationally separate HepG2 and parasite signals, and data analysis.
Materials and reagents
Biological materials
Plasmodium berghei ANKA (676 m1cl1)–infected Anopheles stephensi mosquitos (University of Georgia SporoCore)
HepG2 human hepatoma cell line (ATCC STR profiling-verified)
Reagents
DMEM (Gibco, catalog number: 10313-021)
Pen Strep (Gibco, catalog number: 15140-122)
Heat inactivated fetal bovine serum (FBS) (Gibco, catalog number: 16140-071)
GlutaMAX (Gibco, catalog number: 35050-061)
TrypLE Express (Gibco, catalog number: 12605-028)
Dulbecco’s PBS (DPBS) (Sigma, catalog number: D8537)
PBS (Sigma, catalog number: P3813)
Triton X-100 (Sigma, catalog number: T9284)
Paraformaldehyde (PFA) solution, 4% in PBS (Thermo Scientific, catalog number: 30525-89-4)
Penicillin Streptomycin Neomycin (PSN) (100×) (Gibco, catalog number:15640-055)
Kanamycin sulfate (Corning, catalog number: 30-006-CF)
Amphotericin B (Gibco, catalog number: 15290-026)
Gentamycin (Gibco, catalog number: 15750-060)
DMSO (Sigma, catalog number: D2650)
Anisomycin (EMD Millipore-Sigma, catalog number: 176880)
O-propargyl-puromycin (OPP) (Invitrogen, catalog number: 10459)
O-propargyl-puromycin (OPP) [Vector labs (formerly Click Chemistry Tools), catalog number: CCT-1407-5]
Click-iT® Plus Alexa Fluor® 555 picolyl azide toolkit (Invitrogen, catalog number: C10642)
Click-&-Go® Plus 555 Imaging kit (Vector labs, catalog number: CCT-1317)
Bovine serum albumin (BSA) (Fisher Bioreagents, catalog number: 9048-46-8)
Mouse anti-PbHsp70 primary antibody [13]
Donkey anti-mouse Alexa Fluor® 488 (Invitrogen, catalog number: A21202)
Hoechst 33342 (10 mg/mL) (Thermo Scientific, catalog number: 62249)
70% ethanol
Laboratory supplies
25 cm2 cell culture flask (Corning, catalog number: 430168)
75 cm2 cell culture flask (Corning, catalog number: 430720U)
Cell strainer, 40 μm nylon (Corning, catalog number: 431750)
Cell strainer, 100 µm nylon (Corning, catalog number: 431752)
0.22 µm filters (Millex, catalog number: SLGV004SL)
0.20 µm filters (Corning, catalog number: 431229)
1 mL syringes (BD, catalog number: 309659)
10 mL syringes (BD, catalog number: 302995)
1 mL syringes with hypodermic needle (BD, catalog number: 309626)
50 mL conical tubes (Corning, catalog number: 430828)
24-well cell culture plate (Corning, catalog number: 3524)
12 mm round coverslips, 1.5 thickness (Azer, catalog number: ES0117520)
Fluoromount-G (SouthernBiotech, catalog number: 0100-01)
96-well µClear cell culture plate (Greiner bio-one, catalog number: 655098)
96-well round bottom plate (Corning, catalog number: 3799)
5 mL serological pipettes (Corning, catalog number: 4487)
10 mL serological pipettes (Corning, catalog number: 4488)
25 mL serological pipettes (Corning, catalog number: 4489)
Reagent reservoirs (Eppendorf, catalog number: 022265806)
Live insect forceps (FST, catalog number: 26029-10)
Dumont #5/45 coverslip forceps (FST, catalog number: 11251-33)
6-well plate (Corning, catalog number: 351146)
Neubauer chamber (Brand GMBH, catalog number: 717805)
Pestle (Fisherbrand, catalog number: 12-141-363)
Multichannel racked pipette tips, 1,200 µL (Rainin, catalog number: 17002921)
Multichannel racked pipette tips, 300 µL (Rainin, catalog number: 30389255)
P1000 barrier pipette tips (Thermo Scientific, catalog number: 2079)
Microscope slides 75 × 25 × 1 mm (VWR, catalog number: 16004-368)
1.7 mL snap top tubes (Axygen, catalog number: MCT-175-A)
Kimwipes
Paper towels
Ice bucket
Solutions
Complete DMEM (cDMEM) (see Recipes)
Infection DMEM (iDMEM) (see Recipes)
DMSO control treatment (see Recipes)
Anisomycin control treatment (see Recipes)
0.5% Triton-X (see Recipes)
Blocking solution (see Recipes)
Click-iT® Plus Master Mix (see Recipes)
1° (primary) antibody solution (see Recipes)
2° (secondary) antibody solution (see Recipes)
Recipes
Complete DMEM (cDMEM)
Reagent Final concentration Quantity or Volume DMEM 88% (v/v) 440 mL FBS
Pen Strep
GlutaMAX
10% (v/v)
1% (v/v)
1% (v/v)
50 mL
5 mL
5 mL
Total n/a 500 mL Infection DMEM (iDMEM)
Reagent Final concentration Quantity or Volume cDMEM 97.5% (v/v) 97.5 mL PSN
Kanamycin sulfate
Amphotericin B
Gentamycin
1% (v/v)
1% (v/v)
0.33% (v/v)
0.1% (v/v)
1 mL
1 mL
334 µL
100 µL
Total n/a 100 mL DMSO control treatment
Reagent Final concentration Quantity or Volume iDMEM n/a 1,998 µL DMSO 0.1% (v/v) 2 µL Total n/a 2,000 µL Anisomycin control treatment
Reagent Final concentration Quantity or Volume iDMEM n/a 1,998 µL 10 mM stock of anisomycin 0.1% (v/v) 2 µL Total n/a 2,000 µL 0.5% Triton-X
Reagent Final concentration Quantity or Volume PBS n/a 4,750 µL 10% Triton X-100 0.5% (v/v) 250 µL Total (optional) n/a 5 mL Blocking solution (2% BSA in PBS)
Reagent Final concentration Quantity or Volume BSA 2% (w/v) 1 g PBS n/a 50 mL Total n/a 50 mL Click iT® Plus Master Mix
Note: All reagents should be prepared and stored in accordance with manufacturer’s specifications. Components should be brought to room temperature before making master mix. Each master mix should be prepared fresh and used within 15 min. Each component of the master mix should be added in the order listed. The CuSO4 and copper protectant mix should be combined separately before addition to the master mix; these components are stored separately to allow you to change the concentration of copper catalyst in the reaction master mix by adjusting the ratio of CuSO4 to copper protectant buffer. We have found that a 1:4 ratio of copper–copper protectant provides nearly identical signal compared to those of higher ratios. The total volume reported here is designed for a 96-well plate (32 wells at 27 µL each) with ample dead volume.
Reagent Final concentration Quantity or Volume Molecular grade H2O 78.3% (v/v) 783 µL 10× Click-iT® reaction buffer
500 µM Alexa Fluor® PCA solution
CuSO4-copper protectant mix
1×x Click-iT® buffer additive
8.7% (v/v)
1% (v/v)
2% (v/v)
10% (v/v)
87 µL
10 µL
20 µL
100 µL
Total (optional) n/a 1,000 µL 1° antibody solution
Note: Anti-PbHSP70 working dilution will depend on antibody concentration; the dilution given is for an unpurified batch produced from the 2E6 hybridoma in the Hanson lab. Antibody solutions are filtered using 0.20–0.22 µm filters before use.
Reagent Final concentration Quantity or Volume Blocking solution n/a 1,592 µL Mouse anti-PbHSP70 1:200 (v/v) 8 µL Total (optional) n/a 1,600 µL 2° antibody solution
Note: Antibody solutions are filtered using 0.20–0.22 µm filters before use.
Reagent Final concentration Quantity or Volume Blocking solution n/a 1595.2 µL Donkey anti-mouse Alexa Fluor® 488
Hoechst 33342 (10mg/mL)
1:500 (v/v)
1:1,000 (v/v)
3.2 µL
1.6 µL
Total (optional) n/a 1,600 µL
Equipment
SP8 confocal microscope (Leica, model: TCS SP8)
Dissecting stereoscope (Leica, model: S4E)
Inverted phase contrast microscope (Zeiss, model: Invertoskop 40 C)
Inverted fluorescence microscope (Leica, model: DM IL LED)
Fluorescence light source (Leica, model: EL6000)
Water bath (Thermo, model: 280 series)
CO2 incubator (Thermo, model: Napco Series 8000 WJ)
Biological safety cabinet (LABGARD, model: Class II, Type A2)
Centrifuge with swinging bucket rotor (Thermo, model: Sorval ST 40R, TX-750 rotor)
Titanium liquid removal chambers for swinging bucket rotor centrifuge (custom fabrication by Autotiv)
Small bench centrifuge (Eppendorf, model: 5415D)
Multichannel pipette 300 µL LTS (Rainin, model: E4 XLS)
Multichannel pipette 1200 µL LTS (Rainin, model: E4 XLS)
Software and datasets
LAS AF 3 (Leica)
MatrixScreener (HCS A Developer Full with CAM Leica)
CellProfiler (v2.1.1 rev6c2d896, 7/25/2014), used for image segmentation and feature extraction
CellProfiler (v2.0.11710, 01/24/2014), used for online image segmentation and parasite ID during automated confocal feedback microscopy (ACFM)
KNIME version 4.7.7 and earlier v4 releases (01/20/2022), used for data analysis
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
McLellan, J. L., Garcia-Vilanova, A. and Hanson, K. K. (2024). An Optimized P. berghei Liver Stage–HepG2 Infection Model for Simultaneous Quantitative Bioimaging of Host and Parasite Nascent Proteomes. Bio-protocol 14(5): e4952. DOI: 10.21769/BioProtoc.4952.
分类
微生物学 > 微生物细胞生物学 > 基于细胞的分析方法
细胞生物学 > 细胞成像 > 荧光
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link