- EN - English
- CN - 中文
Development of Recombinase Polymerase Amplification–Lateral Flow Dipstick (RPA–LFD) as a Rapid On-Site Detection Technique for Fusarium oxysporum
重组酶聚合酶扩增-测流层析试纸 (RPA-LFD) 作为尖孢镰刀菌快速现场检测技术的开发
发布: 2024年01月05日第14卷第1期 DOI: 10.21769/BioProtoc.4915 浏览次数: 2008
评审: Shweta PanchalMichael EnosSoumya Moonjely

相关实验方案

用于跨谱应急和现场使用的快速、可持续的热渗透 DNA 提取方案
Stavroula Goudoudaki [...] Yiannis Manoussopoulos
2023年09月05日 1724 阅读

优化高分子量 DNA 提取方法以用于 Magnaporthaceae 及其他禾本科根部真菌的长读长全基因组测序
Michelle J. Grey [...] Mark McMullan
2025年03月20日 3249 阅读
Abstract
Fusarium oxysporum can cause many important plant diseases worldwide, such as crown rot, wilt, and root rot. During the development of strawberry crown rot, this pathogenic fungus spreads from the mother plant to the strawberry seedling through the stolon, with obvious characteristics of latent infection. Therefore, the rapid and timely detection of F. oxysporum can significantly help achieve effective disease management. Here, we present a protocol for the recombinase polymerase amplification– lateral flow dipstick (RPA–LFD) detection technique for the rapid detection of F. oxysporum on strawberry, which only takes half an hour. A significant advantage of our RPA–LFD technique is the elimination of the involvement of professional teams and laboratories, which qualifies it for field detection. We test this protocol directly on plant samples with suspected infection by F. oxysporum in the field and greenhouse. It is worth noting that this protocol can quickly, sensitively, and specifically detect F. oxysporum in soils and plants including strawberry.
Key features
• This protocol is used to detect whether plants such as strawberry are infected with F. oxysporum.
• This protocol has potential for application in portable nucleic acid detection.
• It can complete the detection of samples in the field within 30 min.
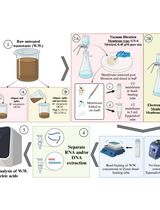
Graphical overview
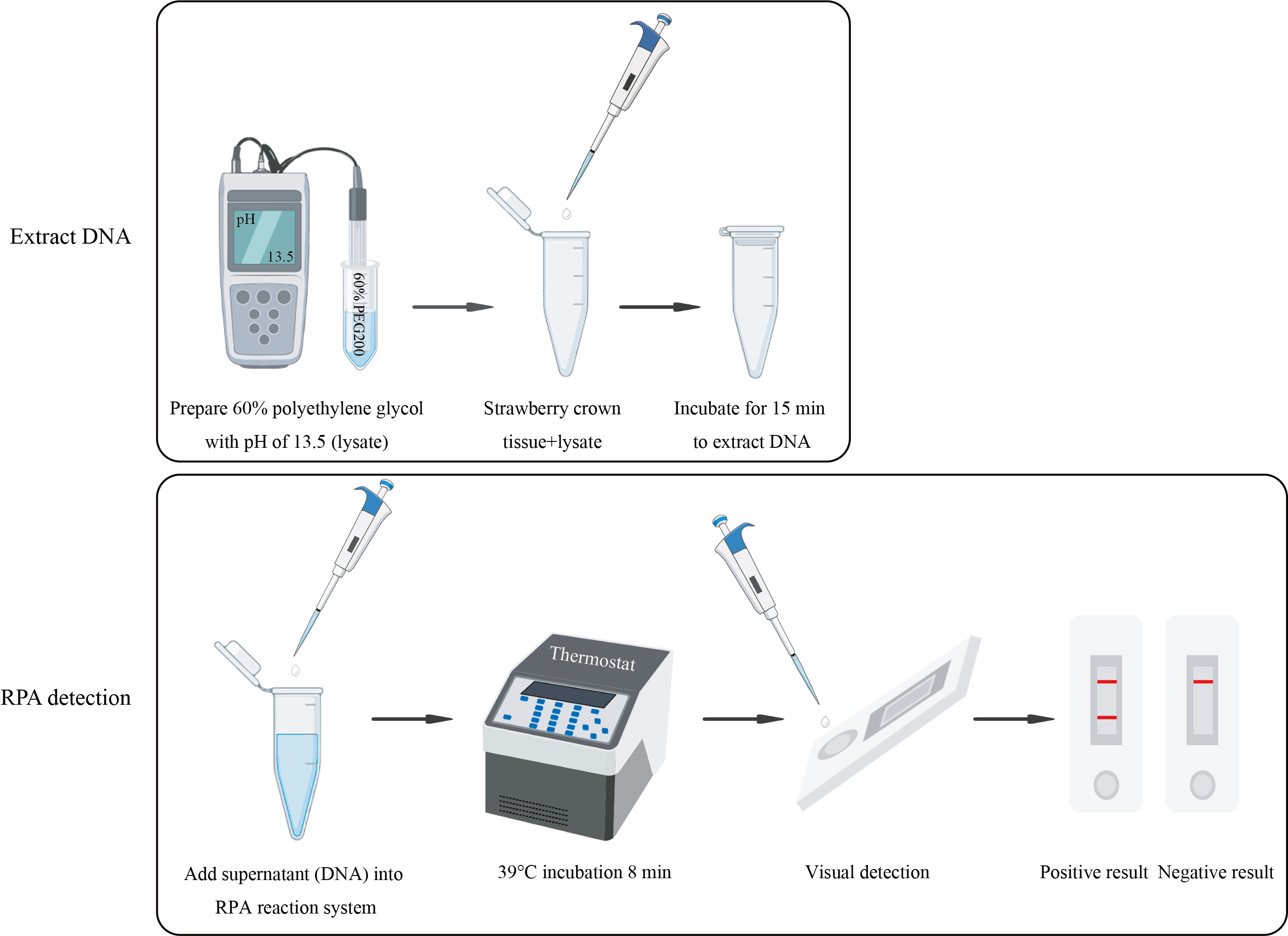
Background
Strawberry crown rot caused by Fusarium oxysporum has seriously affected strawberry yield [1]. The pathogen is transmitted from the mother plant to the strawberry seedling through the stolon [2]. Due to the particularities of pathogen transmission, the control of this disease is difficult. Therefore, rapid and accurate detection of F. oxysporum in the early stages is of utmost importance for preventing and controlling crown rot disease.
Koch’s postulates are the traditional method for pathogen detection through isolation, culture, and pathogenicity testing, being time-consuming and requiring professional training. Molecular detection and identification of the pathogen are highly accurate and sensitive alternative methods for disease diagnosis. Polymerase chain reaction (PCR), quantitative PCR, and other molecular detection techniques have been invented to detect pathogens [3, 4]. However, these techniques require professional experience in performing the tests and expensive equipment, which may not be possible in on-site detection. Altogether, these create a bottleneck for early-disease diagnosis. Fortunately, other molecular biological methods can overcome these limitations, such as the recombinase polymerase amplification (RPA) detection technique [5]. RPA is performed at constant temperature, and its most significant advantage is that the detection results can be visualized by lateral flow dipstick (LFD) [6]. This method is simple to operate and requires little laboratory equipment. Therefore, it has potential for application in portable nucleic acid detection.
Here, we develop and evaluate the RPA–LFD detection technique that can quickly, sensitively, and specifically detect F. oxysporum infection in strawberry [7]. By comparing the genomes of F. oxysporum with other strains common in soil and strawberry plants, we found that the sequence of CYP51C gene was unique and highly variable in Fusarium spp.; therefore, we chose the CYP51C gene to design primers and probes for F. oxysporum detection. We demonstrate how F. oxysporum can be accurately detected on-site. This protocol requires no professional experience, is easy to operate, and the results can be observed within 8 min at 39 °C. This advantage makes it a valuable diagnostic tool, especially in the absence of experienced microscopists. The sensitivity of this protocol is consistent with PCR, and it is accurate enough to detect F. oxysporum. In summary, this protocol is validated on the early field diagnosis of strawberry crown rot. Additionally, it can be applied to detect F. oxysporum in soils and other plants that are damaged by this fungus and can be adjusted to detect other pathogens, only by redesigning specific primers and probes according to the genome sequence of the pathogen to be detected.
Materials and reagents
Biological materials
DNA of F. oxysporum, F. tricinctum, F. proliferatum, F. fujikuroi, F. graminearum, F. solani, F. equiseti, Colletotrichum siamense, C. gloeosporioides, C. aenigma, C. fructicola, Botrytis cinerea, Phoma sp., and Stagonosporopsis sp.
Strawberry plant
Potato
Reagents
KOH (SangonBiotech, catalog number: A610441)
Polyethylene glycol (PEG200) (SangonBiotech, catalog number: A601780-0500)
EDTA disodium salt dihydrate (SangonBiotech, catalog number: A610185-050)
CTAB, cetyltrimethylammonium bromide (SangonBiotech, catalog number: A600108-0500)
Sodium chloride (NaCl) (SangonBiotech, catalog number: A610476-0001)
Tris (SangonBiotech, catalog number: A610195-0500)
Ethanol absolute (SangonBiotech, catalog number: A500737-0500)
Isopropanol (Shanghai Hushi, catalog number: 180109218)
Trichloromethane (Shanghai Hushi, catalog number: 10006818)
Dextrose (SangonBiotech, catalog number: A610219-0500)
Agar (SangonBiotech, catalog number: A505255-025)
Primer and probe:
F91:5′-TCAACTGGCATCGTCAACATCACCGAAGTAA-3′
R91:5′ Biotin-CCAGGCATGACGAAGTTGATAGGTTGAAAGC-3′
Probe:5′ 6-FAM-AGGCTCCCTCCTCGGTAACGAAGTCCGCTCCA/idSp/GTTTGACAGCACATT-3′ C3 Spacer
Solutions
CTAB extracting solution (see Recipes)
70% Ethanol absolute (see Recipes)
60% PEG200 with pH 13.2–13.5 (see Recipes)
2 M KOH (see Recipes)
Potato dextrose agar (PDA) (see Recipes)
Recipes
CTAB extracting solution
*Note: The solute is first mixed and then ddH2O is added to the constant volume of 1,000 mL.
Reagent Final concentration Quantity or Volume CTAB
Tris
EDTA
NaCl
H2O
2%
100 mM
20 mM
1.4 M
n/a
20 g
12.114 g
7.448 mL
81.816 g
to 1,000 mL
Total n/a 1,000 mL 70% ethanol absolute
Reagent Final concentration Quantity or Volume Ethanol absolute
H2O
70% (v/v)
n/a
70 mL
30 mL
Total n/a 100 mL 2 M KOH
*Note: Store at 4 °C.
Reagent Final concentration Quantity or Volume KOH
H2O
2 M
n/a
1.12 g
10 mL
Total n/a 10 mL 60% PEG200 with pH 13.2–13.5
*Note: This solution needs to be prepared and used immediately.
Reagent Final concentration Quantity or Volume KOH (Recipe 3)
PEG200
H2O
n/a
60% (v/v)
n/a
0.33 mL
8 mL
11.67 mL
Total n/a 20 mL Potato dextrose agar (PDA)
*Note: Boil the potato in 1,000 mL of H2O using an induction cooker and pan and filter. Then, add the dextrose and agar to the filtrate.
Reagent Final concentration Quantity or Volume Potato
Dextrose
Agar
H2O
200 g/L
20%
15%
n/a
200 g
20 g
15 g
1,000 mL
Total n/a 1,000 mL
Laboratory supplies
1.5 mL microcentrifuge tubes (Axygen, catalog number: MCT-150-C)
HybriD DNA thermostat rapid amplification kit (APWL, catalog number: WLN8203KIT)
HybriDetect lateral flow dipstick (APWL, catalog number: WLFS8201)
Forceps (Shanghai Leigu, catalog number: W-002903)
Sterile blade (VWR International, catalog number: 55411-050)
Beakers (Shanghai Leigu, catalog number: B-000103)
Stir bars (Shanghai Leigu, catalog number: B-040315)
Graduated cylinders (Shanghai Leigu, catalog number: B-031603)
Glass bottles (Shanghai Leigu, catalog number: B-W00323)
90 mm Petri dish (Nantong Baiyao, catalog number: YJ-90-12g)
Inoculating needle (Shanghai Leigu, catalog number: W-006001)
Forceps (Shanghai Leigu, catalog number: W-002806)
Quartz sand (SangonBiotech, catalog number: A500823-0001)
Equipment
Micropipettes 0.1–2.5 µL, 2–20 µL, 20–200 µL (Eppendorf, model: 3123000217, 3123000233, 3123000250)
Incubator, 25 °C (Shanghai Boxun, model: MJX-250B-Z)
4 °C refrigerator (Haier, model: SC-339JN)
Grinding machine (Shanghai Jingxin, model: JXFSTPRP-24L)
Ultramicro accounting protein analyzer (BioDrop, model: BioDrop µlite+)
Centrifuge (ThermoFisher, model: 75002440)
Thermostat (Hangzhou Aosheng, model: 028-14484-20110001)
Balance (Shanghai Haosheng, model: JA303A)
Induction cooker (Midea, model: RT21E0105)
Pan (Supor, model: ST22P1)
Vortex shaker (SangonBiotech, model: WH-861)
Software and datasets
DNAMAN v6.0.3.99
Snapgene v.5.2.4
Procedure
文章信息
版权信息
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
如何引用
Hu, S., Yu, H. and Zhang, C. (2024). Development of Recombinase Polymerase Amplification–Lateral Flow Dipstick (RPA–LFD) as a Rapid On-Site Detection Technique for Fusarium oxysporum. Bio-protocol 14(1): e4915. DOI: 10.21769/BioProtoc.4915.
分类
微生物学 > 病原体检测
分子生物学 > DNA > DNA 提取
生物科学 > 微生物学
您对这篇实验方法有问题吗?
在此处发布您的问题,我们将邀请本文作者来回答。同时,我们会将您的问题发布到Bio-protocol Exchange,以便寻求社区成员的帮助。
Share
Bluesky
X
Copy link